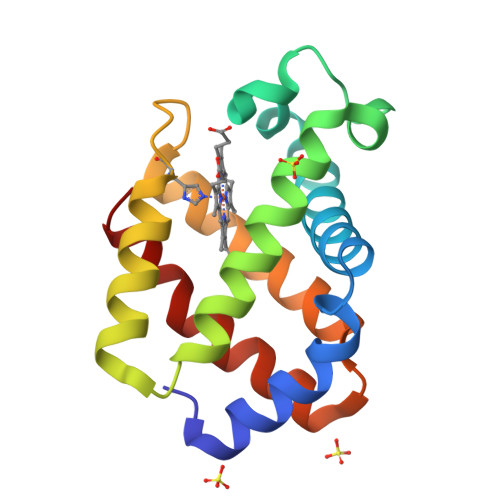
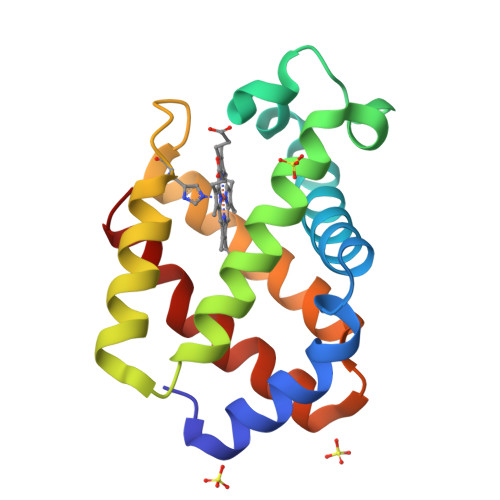
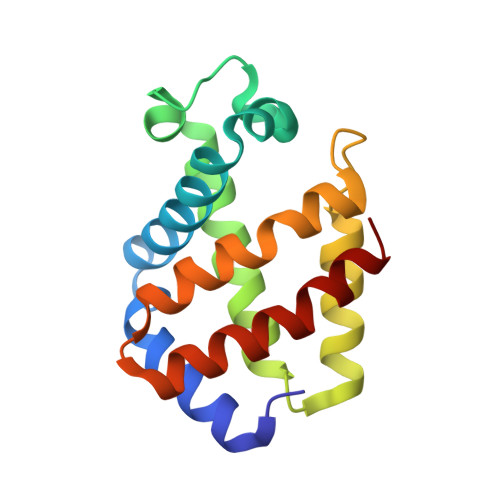
Structural plasticity in the eight-helix fold of a trematode haemoglobin.
Milani, M., Pesce, A., Dewilde, S., Ascenzi, P., Moens, L., Bolognesi, M.(2002) Acta Crystallogr D Biol Crystallogr 58: 719-722
- PubMed: 11914507
- DOI: https://doi.org/10.1107/s0907444902001865
- Primary Citation of Related Structures:
1KFR - PubMed Abstract:
The three-dimensional structure of recombinant haemoglobin from the trematode Paramphistomum epiclitum, displaying the highest oxygen affinity so far observed for (non)vertebrate haemoglobins, has previously been determined at 1.17 A resolution (orthorhombic space group P2(1)2(1)2(1)). In the present communication, the three-dimensional structure of wild-type P. epiclitum haemoglobin is reported at 1.85 A resolution in a monoclinic crystal form (R factor = 16.1%, R(free) = 22.0%). Comparison of P. epiclitum (recombinant versus wild-type ferric Hb) structures in the two crystal forms shows structural differences in the haem proximal and distal sites which have not been reported for other known haemoglobin structures previously.
Organizational Affiliation:
Department of Physics-INFM, Advanced Biotechnology Centre, University of Genova, Largo Rosanna Benzi 10, I-16146 Genova, Italy.