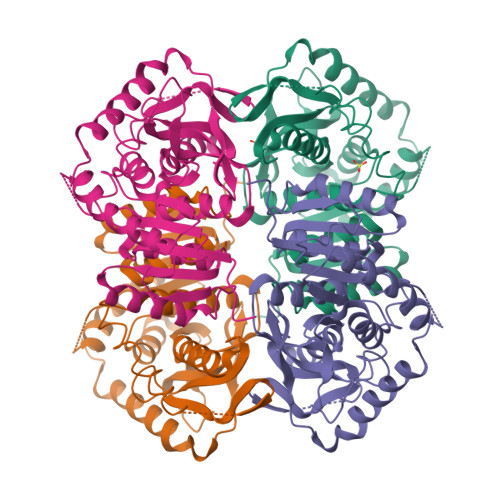
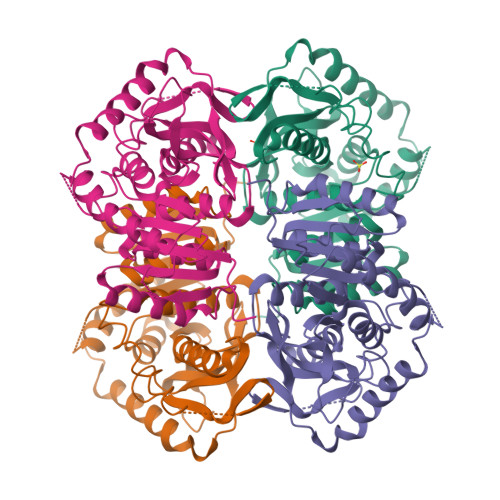
Structure determination and refinement of Bacillus stearothermophilus lactate dehydrogenase.
Piontek, K., Chakrabarti, P., Schar, H.P., Rossmann, M.G., Zuber, H.(1990) Proteins 7: 74-92
- PubMed: 2330370
- DOI: https://doi.org/10.1002/prot.340070108
- Primary Citation of Related Structures:
1LDB, 2LDB - PubMed Abstract:
Structures have been determined of Bacillus stearothermophilus "apo" and holo lactate dehydrogenase. The holo-enzyme had been co-crystallized with the activator fructose 1,6-bisphosphate. The "apo" lactate dehydrogenase structure was solved by use of the known apo-M4 dogfish lactate dehydrogenase molecule as a starting model. Phases were refined and extended from 4 A to 3 A resolution by means of the noncrystallographic molecular 222 symmetry. The R-factor was reduced to 28.7%, using 2.8 A resolution data, in a restrained least-squares refinement in which the molecular symmetry was imposed as a constraint. A low occupancy of coenzyme was found in each of the four subunits of the "apo"-enzyme. Further refinement proceeded with the isomorphous holo-enzyme from Bacillus stearothermophilus. After removing the noncrystallographic constraints, the R-factor dropped from 30.3% to a final value of 26.0% with a 0.019 A and 1.7 degrees r.m.s. deviation from idealized bond lengths and angles, respectively. Two sulfate ions per subunit were included in the final model of the "apo"-form--one at the substrate binding site and one close to the molecular P-axis near the location of the fructose 1,6-bisphosphate activator. The final model of the holo-enzyme incorporated two sulfate ions per subunit, one at the substrate binding site and another close to the R-axis. One nicotinamide adenine dinucleotide coenzyme molecule per subunit and two fructose 1,6-bisphosphate molecules per tetramer were also included. The phosphate positions of fructose 1,6-bisphosphate are close to the sulfate ion near the P-axis in the "apo" model. This structure represents the first reported refined model of an allosteric activated lactate dehydrogenase. The structure of the activated holo-enzyme showed far greater similarity to the ternary complex of dogfish M4 lactate dehydrogenase with nicotinamide adenine dinucleotide and oxamate than to apo-M4 dogfish lactate dehydrogenase. The conformations of nicotinamide adenine dinucleotide and fructose 1,6-bisphosphate were also analyzed.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907.
Organizational Affiliation: