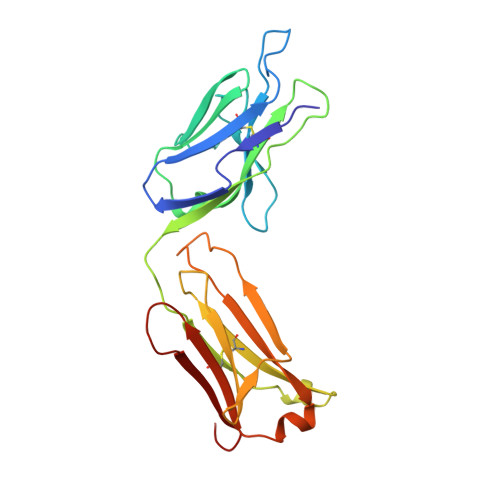
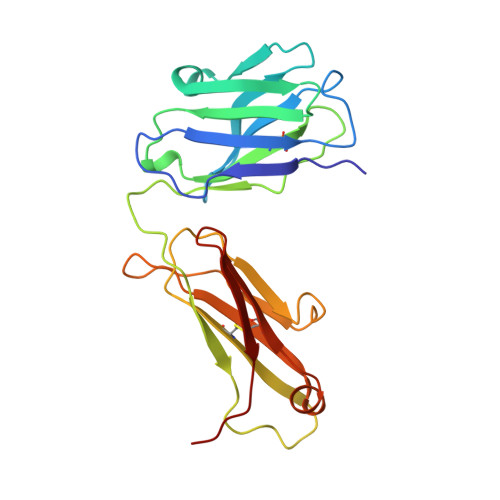
Structure of the Fab fragment from a neutralizing monoclonal antibody directed against an epitope of gp41 from HIV-1.
Davies, C., Beauchamp, J.C., Emery, D., Rawas, A., Muirhead, H.(1997) Acta Crystallogr D Biol Crystallogr 53: 186-194
- PubMed: 15299953
- DOI: https://doi.org/10.1107/S0907444996012048
- Primary Citation of Related Structures:
1NLD - PubMed Abstract:
The structure of a Fab fragment of a monoclonal antibody (1583) that neutralizes a broad range of HIV-1 isolates has been solved by X-ray crystallography. This antibody is directed against a poliovirus/HIV-I chimaera which presents a conserved epitope of the envelope protein gp41. Crystals of 1583 were obtained in the space group P2(1)2(1)2(1) and the structure solved by molecular replacement. The model has been refined against all data in the range 10-2.9 A to a final crystallographic R factor of 0.198. The antigen-binding site features a well defined groove, typical of antibodies that bind to small antigens, created in part by a relatively short CDR H3. The variable regions of 1583 were sequenced and, given the hydrophilic nature of the epitope, revealed a surprising lack of charged residues in the CDR's. However, the antigen-binding cleft is indeed very polar, due in part to the presence of two charged residues that emanate from outside the recognized CDR's.
Organizational Affiliation:
Department of Biochemistry and Molecular Recognition Centre, University of Bristol, England.