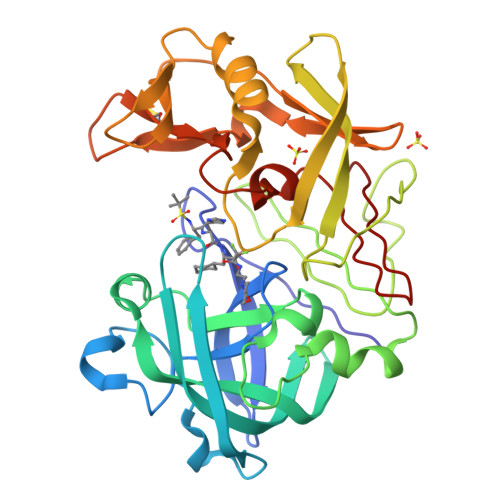
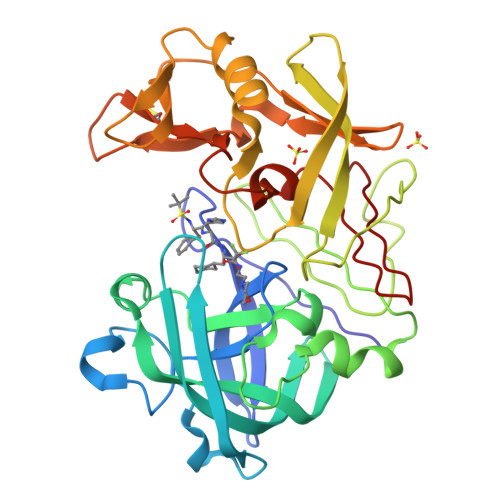
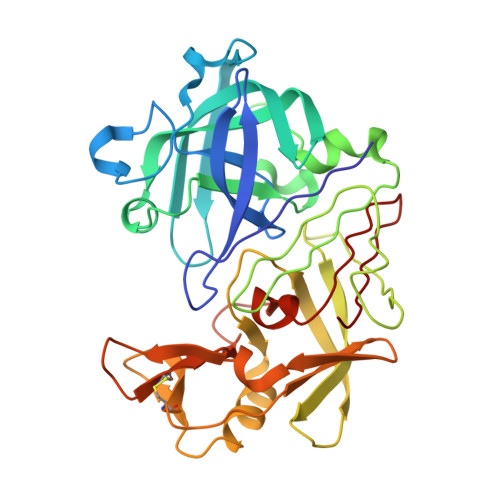
The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Coates, L., Erskine, P.T., Mall, S., Williams, P.A., Gill, R.S., Wood, S.P., Cooper, J.B.(2003) Acta Crystallogr D Biol Crystallogr 59: 978
- PubMed: 12777758
- DOI: https://doi.org/10.1107/s0907444903006267
- Primary Citation of Related Structures:
1OD1 - PubMed Abstract:
The crystal structure of endothiapepsin complexed with the gem-diol inhibitor PD-135,040 has been anisotropically refined to a resolution of 1.37 A. The structure of this inhibitor complex is in agreement with previous structures of endothiapepsin gem-diol inhibitor complexes that have been used to develop proposed catalytic mechanisms. However, the increase in resolution over previous structures confirms the presence of a number of short hydrogen bonds within the active site that are likely to play an important role in the catalytic mechanism. The presence of low-barrier hydrogen bonds was indicated in a previous one-dimensional H NMR spectrum.
Organizational Affiliation:
School of Biological Sciences, University of Southampton, Bassett Crescent East, Southampton SO16 7PX, England. leightonc@bigfoot.com