Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
Jonsson, S., Ricagno, S., Lindqvist, Y., Richards, N.G.(2004) J Biol Chem 279: 36003-36012
- PubMed: 15213226
- DOI: https://doi.org/10.1074/jbc.M404873200
- Primary Citation of Related Structures:
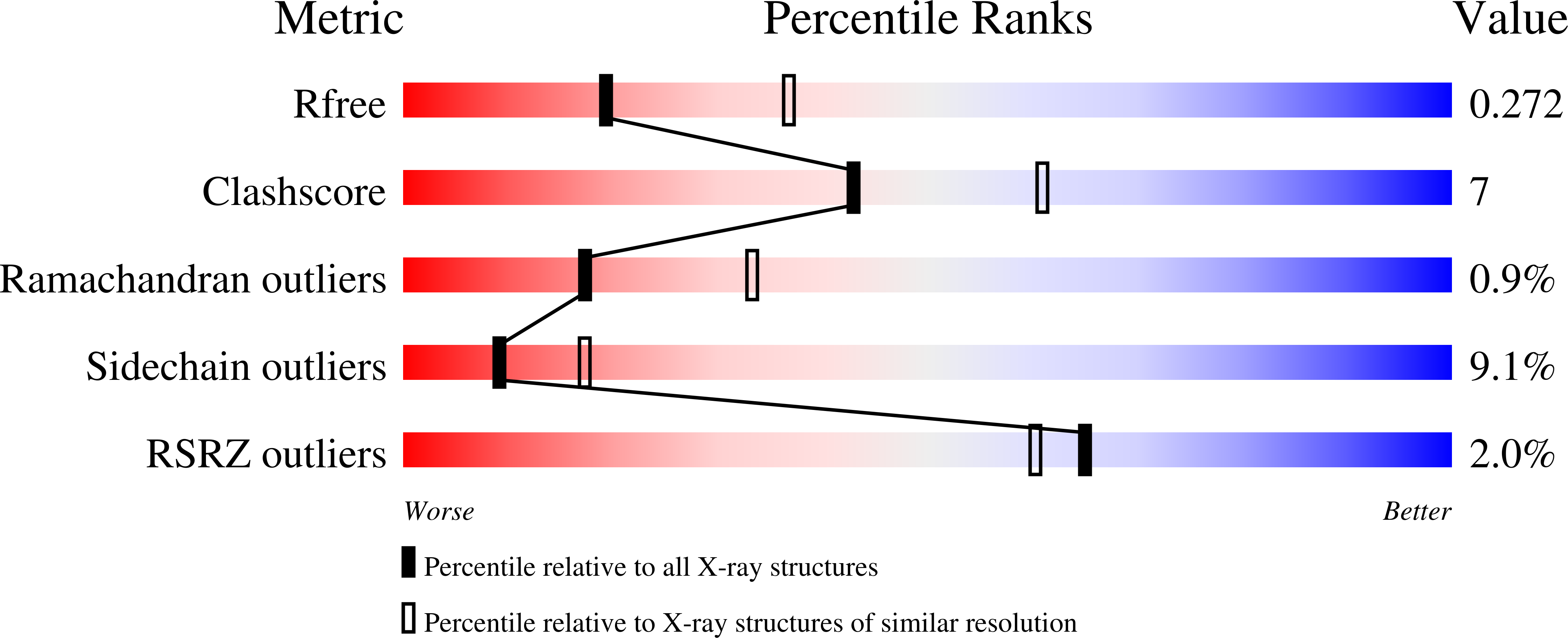
1T3Z, 1T4C, 1VGQ, 1VGR - PubMed Abstract:
Oxalobacter formigenes is an obligate anaerobe that colonizes the human gastrointestinal tract and employs oxalate breakdown to generate ATP in a novel process involving the interplay of two coupled enzymes and a membrane-bound oxalate:formate antiporter. Formyl-CoA transferase is a critical enzyme in oxalate-dependent ATP synthesis and is the first Class III CoA-transferase for which a high resolution, three-dimensional structure has been determined (Ricagno, S., Jonsson, S., Richards, N., and Lindqvist, Y. (2003) EMBO J. 22, 3210-3219). We now report the first detailed kinetic characterizations of recombinant, wild type formyl-CoA transferase and a number of site-specific mutants, which suggest that catalysis proceeds via a series of anhydride intermediates. Further evidence for this mechanistic proposal is provided by the x-ray crystallographic observation of an acylenzyme intermediate that is formed when formyl-CoA transferase is incubated with oxalyl-CoA. The catalytic mechanism of formyl-CoA transferase is therefore established and is almost certainly employed by all other members of the Class III CoA-transferase family.
Organizational Affiliation:
Department of Chemistry, University of Florida, Gainesville, Florida 32611-7200, USA.