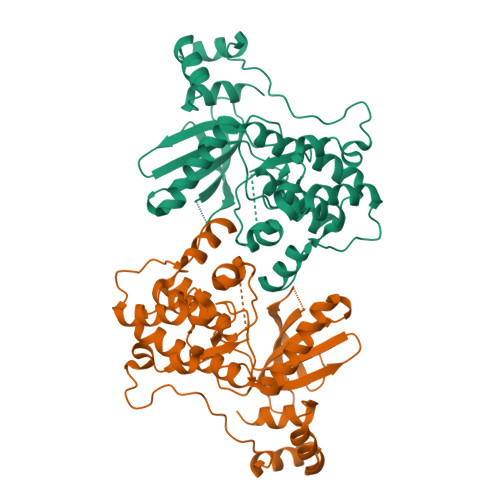
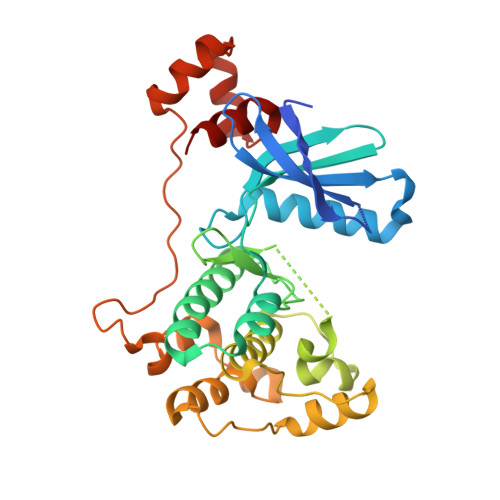
Structure of the catalytic and ubiquitin-associated domains of the protein kinase MARK/Par-1.
Panneerselvam, S., Marx, A., Mandelkow, E.M., Mandelkow, E.(2006) Structure 14: 173-183
- PubMed: 16472737
- DOI: https://doi.org/10.1016/j.str.2005.09.022
- Primary Citation of Related Structures:
1Y8G, 1ZMU, 1ZMV, 1ZMW - PubMed Abstract:
The Ser/Thr kinase MARK2 phosphorylates tau protein at sites that cause detachment from microtubules in Alzheimer neurofibrillary degeneration. Homologs of MARK2 include Par-1 in C. elegans and Drosophila, which generates embryonic polarity. We report the X-ray structure of the catalytic and ubiquitin-associated domains (UBA) of human MARK2. The activity was altered by mutations in the ATP binding site and/or activation loop. The catalytic domain shows the small and large lobes typical of kinases. The substrate cleft is in an inactive, open conformation in the inactivated and the wild-type structure. The UBA domain is attached via a taut linker to the large lobe of the kinase domain and leans against a hydrophobic patch on the small lobe. The UBA structure is unusual because the orientation of its third helix is inverted, relative to previous structures. Possible implications of the structure for the regulation of kinase activity are discussed.
Organizational Affiliation:
Max Planck Unit for Structural Molecular Biology, Notkestrasse 85, 22607 Hamburg, Germany.