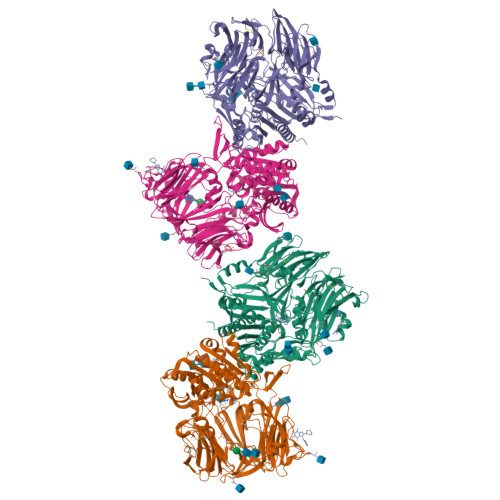
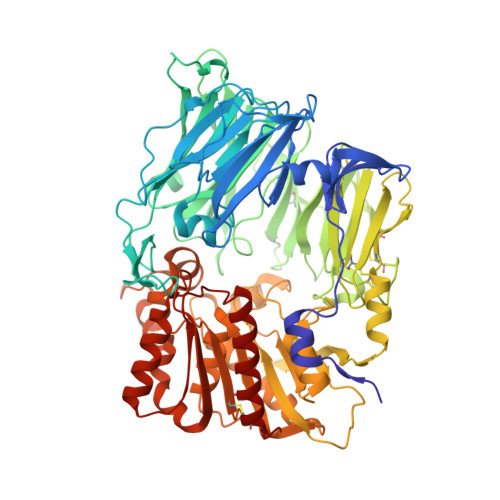
Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
Engel, M., Hoffmann, T., Manhart, S., Heiser, U., Chambre, S., Huber, R., Demuth, H.U., Bode, W.(2006) J Mol Biology 355: 768-783
- PubMed: 16330047
- DOI: https://doi.org/10.1016/j.jmb.2005.11.014
- Primary Citation of Related Structures:
2AJ8, 2AJB, 2AJC, 2AJD - PubMed Abstract:
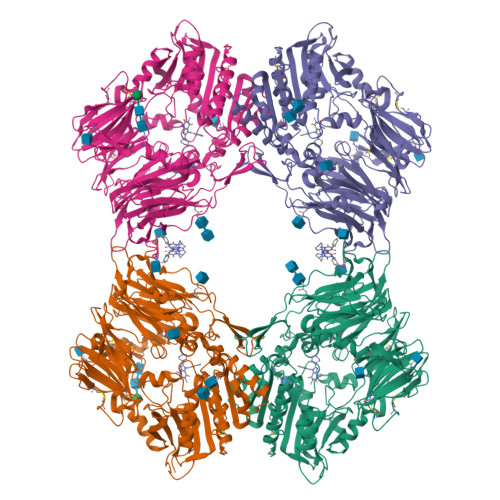
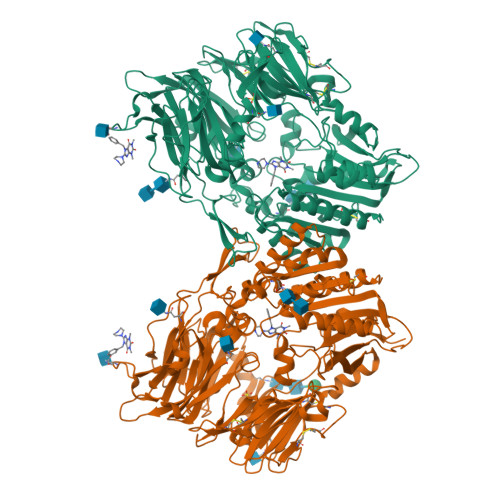
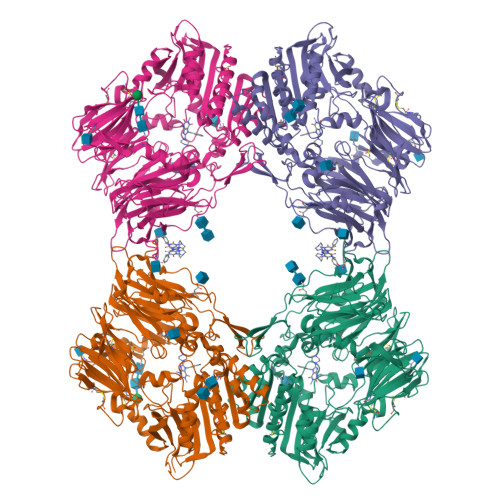
Dipeptidyl peptidase IV (DPIV) is an alpha,beta-hydrolase-like serine exopeptidase, which removes dipeptides, preferentially with a C-terminal l-Pro residue, from the N terminus of longer peptide substrates. Previously, we determined the tetrameric 1.8A crystal structure of native porcine DPIV. Each monomer is composed of a beta-propeller and a catalytic domain, which together embrace an internal cavity housing the active centre. This cavity is connected to the bulk solvent by a "propeller opening" and a "side opening". Here, we analyse DPIV complexes with a t-butyl-Gly-Pro-Ile tripeptide, Pro-boroPro, a piperazine purine compound, and aminoethyl phenyl sulfonylfluoride. The latter two compounds bind to the active-site groove in a compact and a quite bulky manner, respectively, causing considerable shifts of the catalytic Ser630 side-chain and of the Tyr547 phenolic group, which forms the oxyanion hole. The tripeptide, mimicking a peptide substrate, is clamped to the active site through tight interactions via its N-terminal alpha-ammonium group, the P2 carbonyl group, the P1-l-Pro side-chain, the C-terminal carboxylate group, and the stable orthoacid ester amide formed between the scissile peptide carbonyl group and Ser630 O(gamma). This stable trapping of the tripeptide could be due to stabilization of the protonated His740 imidazolium cation by the adjacent negatively charged C-terminal carboxylate group, preventing proton transfer to the leaving group nitrogen atom. Docking experiments with the compact rigid 58 residue protein aprotinin, which had been shown to be processed by DPIV, indicate that the Arg1-Pro2 N terminus can access the DPIV active site only upon widening of its side openings, probably by separation of the first and the last propeller blades, and/or of the catalytic and the propeller domain.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, AG Proteinaseforschung, Am Klopferspitz 18, D-82152 Martinsried, Germany.