The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
Brondijk, T.H., van Boxel, G.I., Mather, O.C., Quirk, P.G., White, S.A., Jackson, J.B.(2006) J Biological Chem 281: 13345-13354
- PubMed: 16533815
- DOI: https://doi.org/10.1074/jbc.M513230200
- Primary Citation of Related Structures:
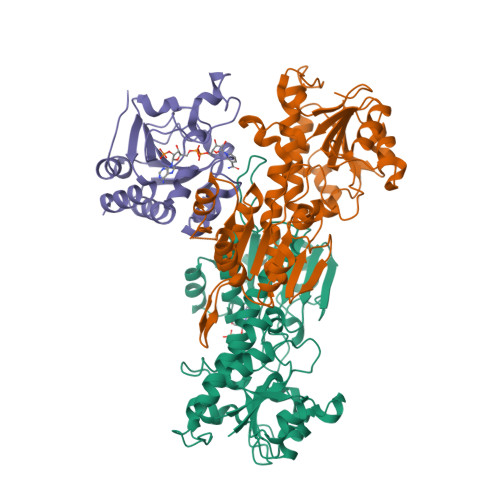
2FR8, 2FRD, 2FSV - PubMed Abstract:
Transhydrogenase couples proton translocation across a membrane to hydride transfer between NADH and NADP+. Previous x-ray structures of complexes of the nucleotide-binding components of transhydrogenase ("dI2dIII1" complexes) indicate that the dihydronicotinamide ring of NADH can move from a distal position relative to the nicotinamide ring of NADP+ to a proximal position. The movement might be responsible for gating hydride transfer during proton translocation. We have mutated three invariant amino acids, Arg-127, Asp-135, and Ser-138, in the NAD(H)-binding site of Rhodospirillum rubrum transhydrogenase. In each mutant, turnover by the intact enzyme is strongly inhibited. Stopped-flow experiments using dI2dIII1 complexes show that inhibition results from a block in the steps associated with hydride transfer. Mutation of Asp-135 and Ser-138 had no effect on the binding affinity of either NAD+ or NADH, but mutation of Arg-127 led to much weaker binding of NADH and slightly weaker binding of NAD+. X-ray structures of dI2dIII1 complexes carrying the mutations showed that their effects were restricted to the locality of the bound NAD(H). The results are consistent with the suggestion that in wild-type protein movement of the Arg-127 side chain, and its hydrogen bonding to Asp-135 and Ser-138, stabilizes the dihydronicotinamide of NADH in the proximal position for hydride transfer.
Organizational Affiliation:
School of Biosciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, United Kingdom.