Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Brugel, T.A., Maier, J.A., Clark, M.P., Sabat, M., Golebiowski, A., Bookland, R.G., Laufersweiler, M.J., Laughlin, S.K., Vanrens, J.C., De, B., Hsieh, L.C., Mekel, M.J., Janusz, M.J.(2006) Bioorg Med Chem Lett 16: 3510-3513
- PubMed: 16632356
- DOI: https://doi.org/10.1016/j.bmcl.2006.03.095
- Primary Citation of Related Structures:
2GHL, 2GHM - PubMed Abstract:
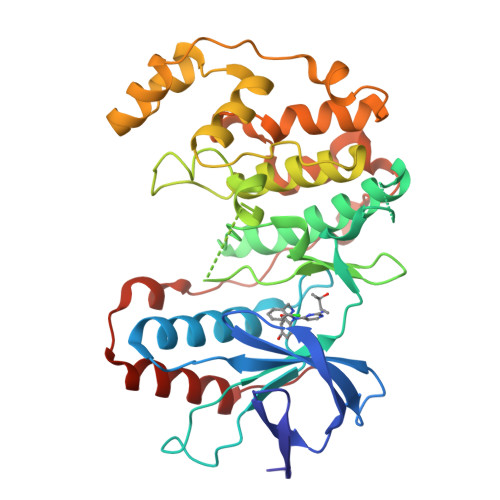
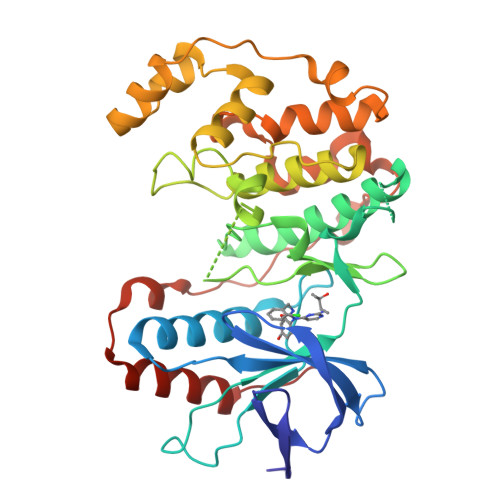
A new class of tumor necrosis factor alpha (TNF-alpha) synthesis inhibitors based on an N-2,4-pyrimidine-N-phenyl-N'-phenyl urea scaffold is described. Many of these compounds showed low-nanomolar activity against lipopolysaccharide stimulated TNF-alpha production. X-ray co-crystallization studies with mutated p38alpha showed that these trisubstituted ureas interact with the ATP-binding pocket in a pseudo-bicyclic conformation brought about by the presence of an intramolecular hydrogen bonding interaction.
Organizational Affiliation:
Procter and Gamble Pharmaceuticals, Health Care Research Center, 8700 Mason-Montgomery Rd., Mason, OH 45040, USA. brugel.ta@pg.com