Structure-reactivity correlations and kinetic isotope effects in aromatic amine dehydrogenase
Hothi, P., Roujeinikova, A., Sutcliffe, M.J., Cullis, P., Leys, D., Scrutton, N.S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aromatic amine dehydrogenase | A [auth D], B [auth H] | 135 | Alcaligenes faecalis | Mutation(s): 0 EC: 1.4.99.4 (PDB Primary Data), 1.4.9.2 (UniProt) | 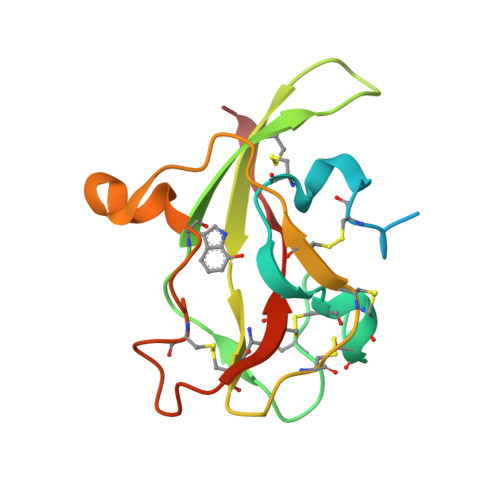 |
UniProt | |||||
Find proteins for P84887 (Alcaligenes faecalis) Explore P84887 Go to UniProtKB: P84887 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84887 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Aromatic amine dehydrogenase | C [auth A], D [auth B] | 362 | Alcaligenes faecalis | Mutation(s): 0 EC: 1.4.99.4 (PDB Primary Data), 1.4.9.2 (UniProt) | 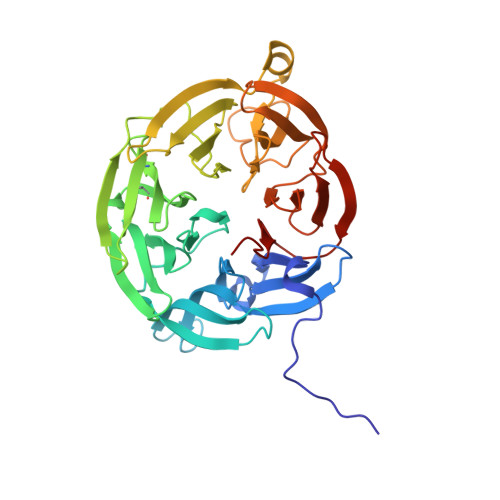 |
UniProt | |||||
Find proteins for P84888 (Alcaligenes faecalis) Explore P84888 Go to UniProtKB: P84888 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P84888 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEA Query on PEA | E [auth D], F [auth H] | 2-PHENYLETHYLAMINE C8 H12 N BHHGXPLMPWCGHP-UHFFFAOYSA-O |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| TRQ Query on TRQ | A [auth D], B [auth H] | L-PEPTIDE LINKING | C11 H10 N2 O4 |  | TRP |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 70.398 | α = 90 |
| b = 89.109 | β = 90.47 |
| c = 80.082 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALA | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| MOSFLM | data reduction |