Inhibitor-complexed Structures of the Cytochrome bc1 from the Photosynthetic Bacterium Rhodobacter sphaeroides.
Esser, L., Elberry, M., Zhou, F., Yu, C.A., Yu, L., Xia, D.(2008) J Biol Chem 283: 2846-2857
- PubMed: 18039651
- DOI: https://doi.org/10.1074/jbc.M708608200
- Primary Citation of Related Structures:
2QJK, 2QJP, 2QJY - PubMed Abstract:
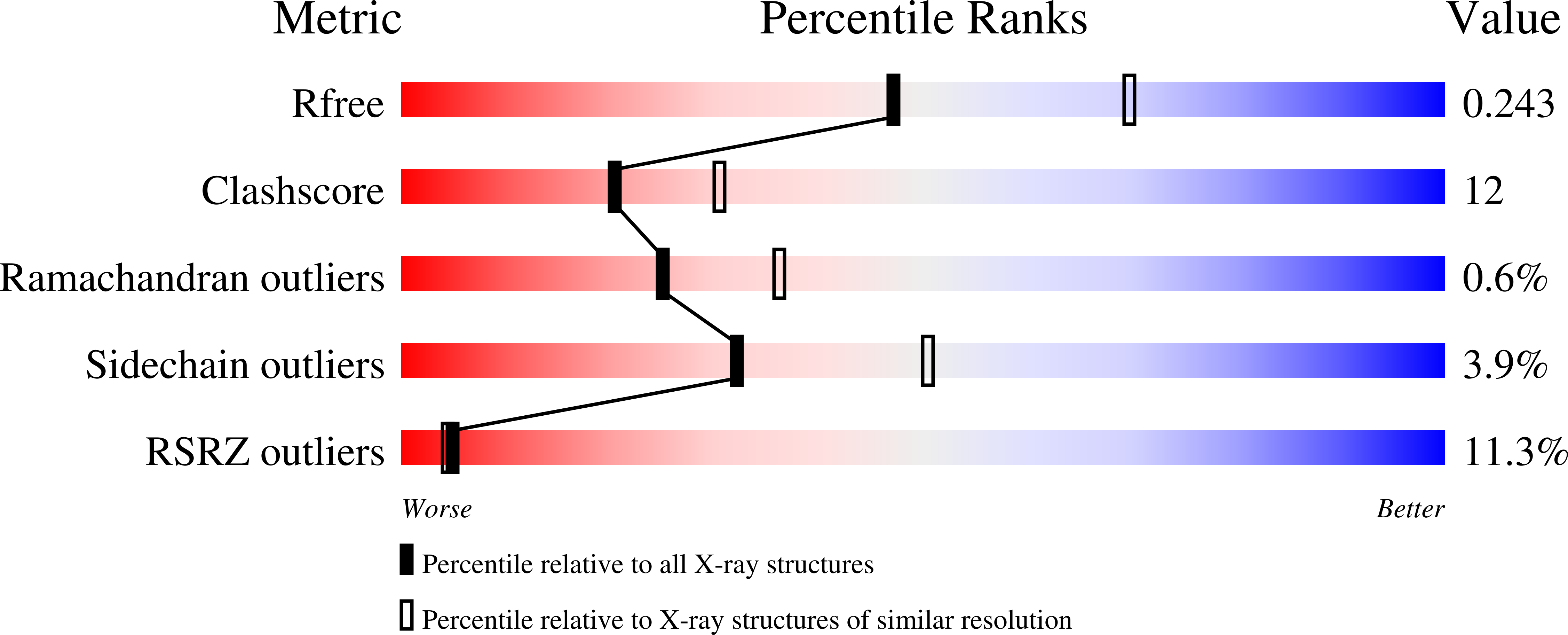
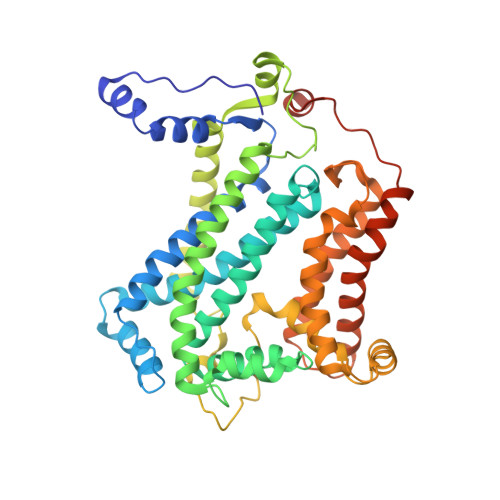
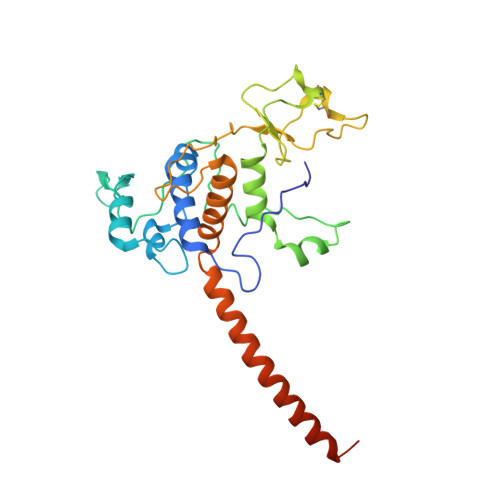
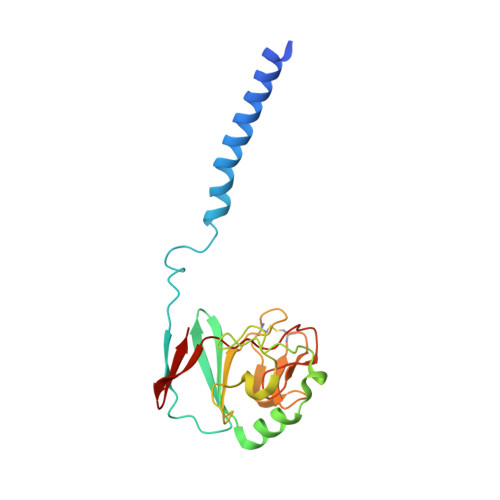
The cytochrome bc(1) complex (bc(1)) is a major contributor to the proton motive force across the membrane by coupling electron transfer to proton translocation. The crystal structures of wild type and mutant bc(1) complexes from the photosynthetic purple bacterium Rhodobacter sphaeroides (Rsbc(1)), stabilized with the quinol oxidation (Q(P)) site inhibitor stigmatellin alone or in combination with the quinone reduction (Q(N)) site inhibitor antimycin, were determined. The high quality electron density permitted assignments of a new metal-binding site to the cytochrome c(1) subunit and a number of lipid and detergent molecules. Structural differences between Rsbc(1) and its mitochondrial counterparts are mostly extra membranous and provide a basis for understanding the function of the predominantly longer sequences in the bacterial subunits. Functional implications for the bc(1) complex are derived from analyses of 10 independent molecules in various crystal forms and from comparisons with mitochondrial complexes.
Organizational Affiliation:
Laboratory of Cell Biology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.