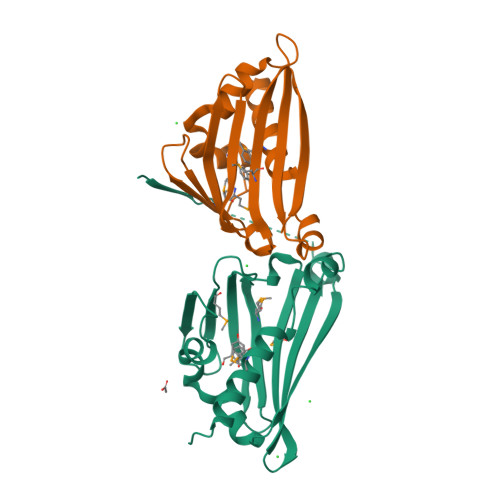
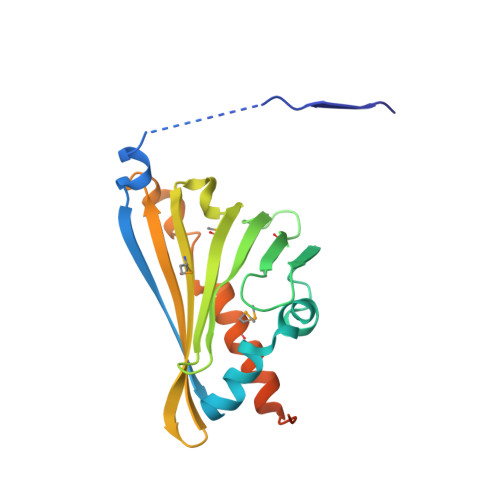
Structural Basis of Enzymatic S-Norcoclaurine Biosynthesis.
Ilari, A., Franceschini, S., Bonamore, A., Arenghi, F., Botta, B., Macone, A., Pasquo, A., Bellucci, L., Boffi, A.(2009) J Biological Chem 284: 897
- PubMed: 19004827
- DOI: https://doi.org/10.1074/jbc.M803738200
- Primary Citation of Related Structures:
2VNE, 2VQ5 - PubMed Abstract:
The enzyme norcoclaurine synthase (NCS) catalyzes the stereospecific Pictet-Spengler cyclization between dopamine and 4-hydroxyphenylacetaldehyde, the key step in the benzylisoquinoline alkaloid biosynthetic pathway. The crystallographic structure of norcoclaurine synthase from Thalictrum flavum in its complex with dopamine substrate and the nonreactive substrate analogue 4-hydroxybenzaldehyde has been solved at 2.1A resolution. NCS shares no common features with the functionally correlated "Pictet-Spenglerases" that catalyze the first step of the indole alkaloids pathways and conforms to the overall fold of the Bet v1-like protein. The active site of NCS is located within a 20-A-long catalytic tunnel and is shaped by the side chains of a tyrosine, a lysine, an aspartic, and a glutamic acid. The geometry of the amino acid side chains with respect to the substrates reveals the structural determinants that govern the mechanism of the stereoselective Pictet-Spengler cyclization, thus establishing an excellent foundation for the understanding of the finer details of the catalytic process. Site-directed mutations of the relevant residues confirm the assignment based on crystallographic findings.
Organizational Affiliation:
Dipartimento di Scienze Biochimiche and Istituto di Biologia e Patologia Molecolari, CNR, Sapienza University, Piazzale Aldo Moro 5, 00185 Roma, Italy.