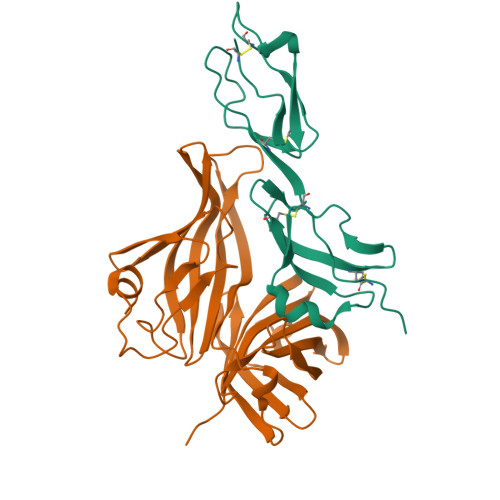
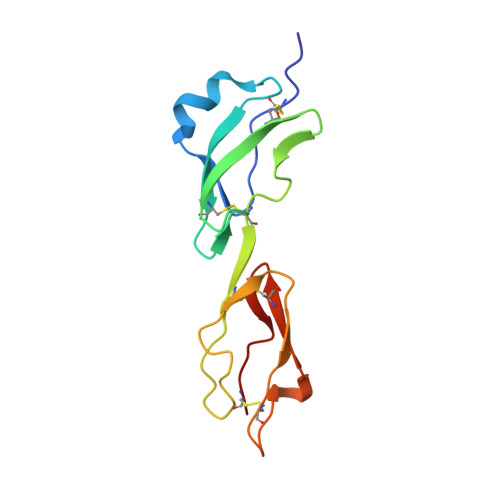
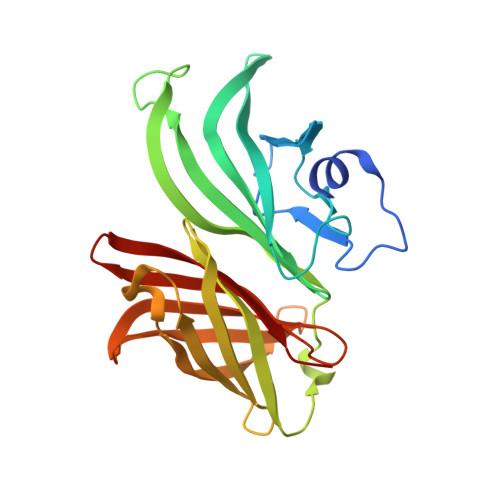
Neisseria Meningitidis Recruits Factor H Using Protein Mimicry of Host Carbohydrates.
Schneider, M.C., Prosser, B.E., Caesar, J.J.E., Kugelberg, E., Li, S., Zhang, Q., Quoraishi, S., Lovett, J.E., Deane, J.E., Sim, R.B., Roversi, P., Johnson, S., Tang, C.M., Lea, S.M.(2009) Nature 458: 890
- PubMed: 19225461
- DOI: https://doi.org/10.1038/nature07769
- Primary Citation of Related Structures:
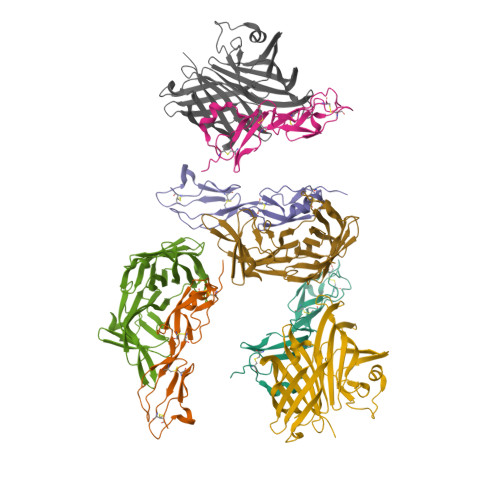
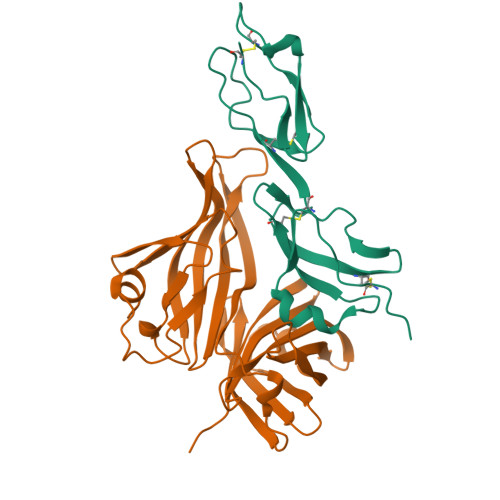
2W80, 2W81 - PubMed Abstract:
The complement system is an essential component of the innate and acquired immune system, and consists of a series of proteolytic cascades that are initiated by the presence of microorganisms. In health, activation of complement is precisely controlled through membrane-bound and soluble plasma-regulatory proteins including complement factor H (fH; ref. 2), a 155 kDa protein composed of 20 domains (termed complement control protein repeats). Many pathogens have evolved the ability to avoid immune-killing by recruiting host complement regulators and several pathogens have adapted to avoid complement-mediated killing by sequestering fH to their surface. Here we present the structure of a complement regulator in complex with its pathogen surface-protein ligand. This reveals how the important human pathogen Neisseria meningitidis subverts immune responses by mimicking the host, using protein instead of charged-carbohydrate chemistry to recruit the host complement regulator, fH. The structure also indicates the molecular basis of the host-specificity of the interaction between fH and the meningococcus, and informs attempts to develop novel therapeutics and vaccines.
Organizational Affiliation:
Centre for Molecular Microbiology and Infection, Imperial College, London SW7 2AZ, UK.