Globin-Like Proteins in Caenorhabditis Elegans: In Vivo Localization, Ligand Binding and Structural Properties.
Geuens, E., Hoogewijs, D., Nardini, M., Vinck, E., Pesce, A., Kiger, L., Fago, A., Tilleman, L., De Henau, S., Marden, M., Weber, R.E., Van Doorslaer, S., Vanfleteren, J., Moens, L., Bolognesi, M., De Wilde, S.(2010) BMC Biochem 11: 17
- PubMed: 20361867
- DOI: https://doi.org/10.1186/1471-2091-11-17
- Primary Citation of Related Structures:
2WTG, 2WTH - PubMed Abstract:
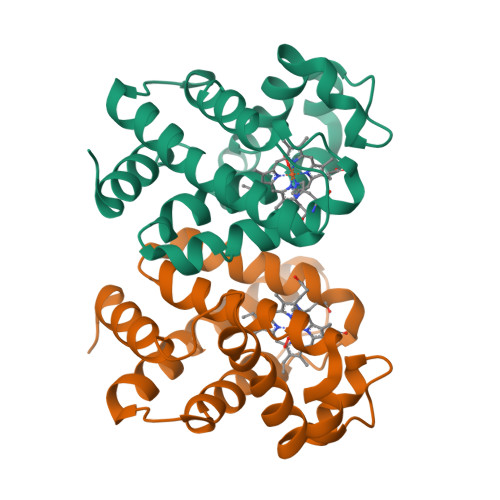
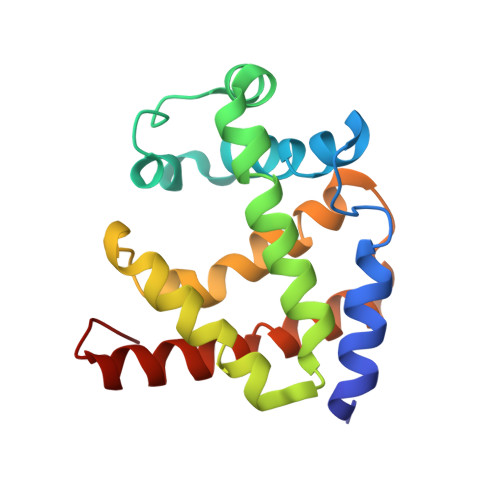
The genome of the nematode Caenorhabditis elegans contains more than 30 putative globin genes that all are transcribed. Although their translated amino acid sequences fit the globin fold, a variety of amino-acid substitutions and extensions generate a wide structural diversity among the putative globins. No information is available on the physicochemical properties and the in vivo expression. We expressed the globins in a bacterial system, characterized the purified proteins by optical and resonance Raman spectroscopy, measured the kinetics and equilibria of O2 binding and determined the crystal structure of GLB-1* (CysGH2 --> Ser mutant). Furthermore, we studied the expression patterns of glb-1 (ZK637.13) and glb-26 (T22C1.2) in the worms using green fluorescent protein technology and measured alterations of their transcript abundances under hypoxic conditions.GLB-1* displays the classical three-over-three alpha-helical sandwich of vertebrate globins, assembled in a homodimer associated through facing E- and F-helices. Within the heme pocket the dioxygen molecule is stabilized by a hydrogen bonded network including TyrB10 and GlnE7.GLB-1 exhibits high ligand affinity, which is, however, lower than in other globins with the same distal TyrB10-GlnE7 amino-acid pair. In the absence of external ligands, the heme ferrous iron of GLB-26 is strongly hexacoordinated with HisE7, which could explain its extremely low affinity for CO. This globin oxidizes instantly to the ferric form in the presence of oxygen and is therefore incapable of reversible oxygen binding. The presented data indicate that GLB-1 and GLB-26 belong to two functionally-different globin classes.
Organizational Affiliation:
Department of Biomedical Sciences, University of Antwerp, Universiteitsplein 1, B-2610 Antwerp, Belgium.