Discovery of novel 5-benzylidenerhodanine and 5-benzylidenethiazolidine-2,4-dione inhibitors of MurD ligase.
Zidar, N., Tomasic, T., Sink, R., Rupnik, V., Kovac, A., Turk, S., Patin, D., Blanot, D., Contreras Martel, C., Dessen, A., Muller Premru, M., Zega, A., Gobec, S., Peterlin Masic, L., Kikelj, D.(2010) J Med Chem 53: 6584-6594
- PubMed: 20804196
- DOI: https://doi.org/10.1021/jm100285g
- Primary Citation of Related Structures:
2WJP, 2X5O - PubMed Abstract:
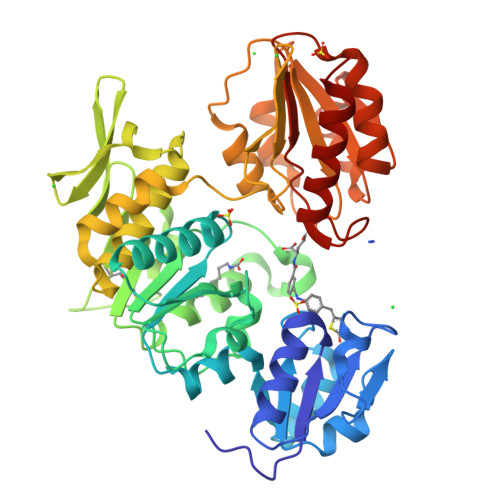
We have designed, synthesized, and evaluated 5-benzylidenerhodanine- and 5-benzylidenethiazolidine-2,4-dione-based compounds as inhibitors of bacterial enzyme MurD with E. coli IC(50) in the range 45-206 μM. The high-resolution crystal structure of MurD in complex with (R,Z)-2-(3-[{4-([2,4-dioxothiazolidin-5-ylidene]methyl)phenylamino}methyl)benzamido)pentanedioic acid [(R)-32] revealed details of the binding mode of the inhibitor within the active site and provides a good foundation for structure-based design of a novel generation of MurD inhibitors.
Organizational Affiliation:
Faculty of Pharmacy, University of Ljubljana, 1000 Ljubljana, Slovenia.