Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors
Lee, C.C., Kuo, C.J., Hsu, M.F., Liang, P.H., Fang, J.M., Shie, J.J., Wang, A.H.(2007) FEBS Lett 581: 5454-5458
- PubMed: 17981158
- DOI: https://doi.org/10.1016/j.febslet.2007.10.048
- Primary Citation of Related Structures:
2Z94, 2Z9G, 2Z9J, 2Z9K, 2Z9L - PubMed Abstract:
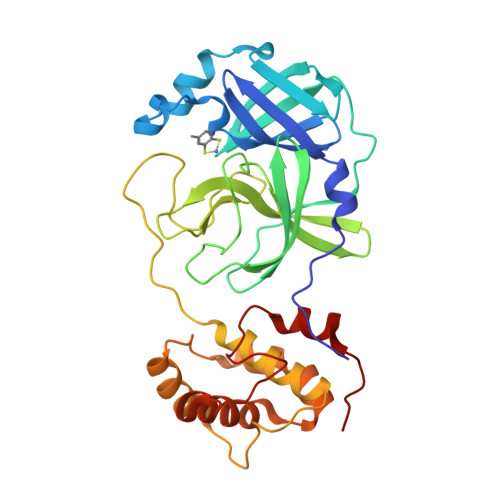
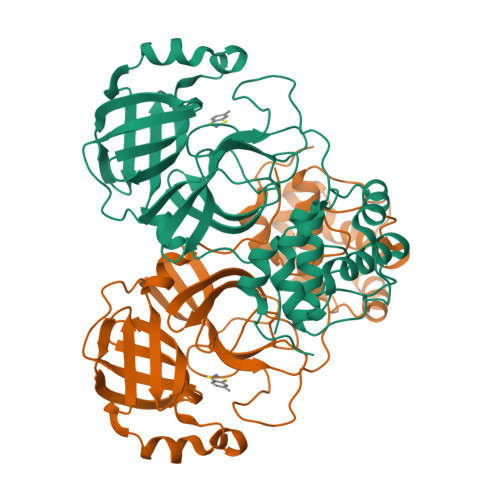
Five active metal-conjugated inhibitors (PMA, TDT, EPDTC, JMF1586 and JMF1600) bound with the 3C-like protease of severe acute respiratory syndrome (SARS)-associated coronavirus were analyzed crystallographically. The complex structures reveal two major inhibition modes: Hg(2+)-PMA is coordinated to C(44), M(49) and Y(54) with a square planar geometry at the S3 pocket, whereas each Zn(2+) of the four zinc-inhibitors is tetrahedrally coordinated to the H(41)-C(145) catalytic dyad. For anti-SARS drug design, this Zn(2+)-centered coordination pattern would serve as a starting platform for inhibitor optimization.
Organizational Affiliation:
Structural Biology Program, Institute of Biochemistry and Molecular Biology, National Yang-Ming University, Taipei 11221, Taiwan.