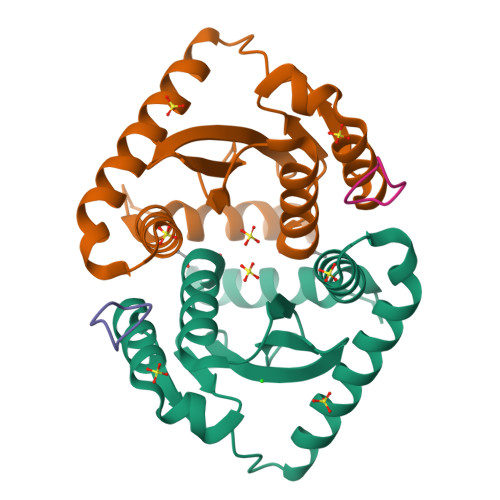
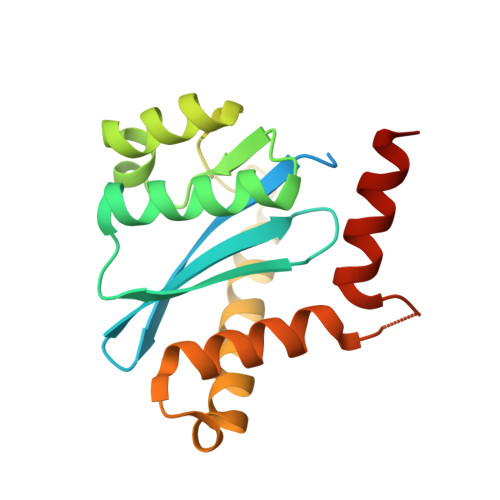
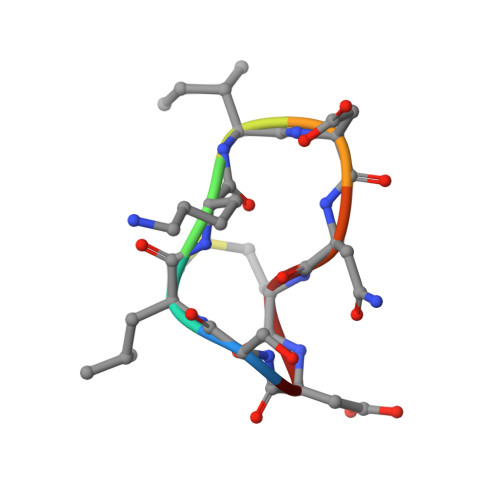
Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Rhodes, D.I., Peat, T.S., Vandegraaff, N., Jeevarajah, D., Newman, J., Martyn, J., Coates, J.A., Ede, N.J., Rea, P., Deadman, J.J.(2011) Chembiochem 12: 2311-2315
- PubMed: 21850718
- DOI: https://doi.org/10.1002/cbic.201100350
- Primary Citation of Related Structures:
3AV9, 3AVA, 3AVB, 3AVC, 3AVF, 3AVG, 3AVH, 3AVI, 3AVJ, 3AVK, 3AVL, 3AVM, 3AVN - PubMed Abstract:
An optimised method of solution cyclisation gave us access to a series of peptides including SLKIDNLD (2). We investigated the crystallographic complexes of the HIV integrase (HIV-IN) catalytic core domain with 13 of the peptides and identified multiple interactions at the binding site, including hydrogen bonds with residues Thr125 and Gln95, that have not previously been described as being accessible within the binding site. We show that the peptides inhibit the interaction of lens epithelium-derived growth factor (LEDGF) with HIV-IN in a proximity AlphaScreen assay and in an assay for the LEDGF enhancement of HIV-IN strand transfer. The interactions identified represent a potential framework for the development of new HIV-IN inhibitors.
Organizational Affiliation:
Avexa Ltd, 576 Swan Street, Melbourne 3121, Australia.