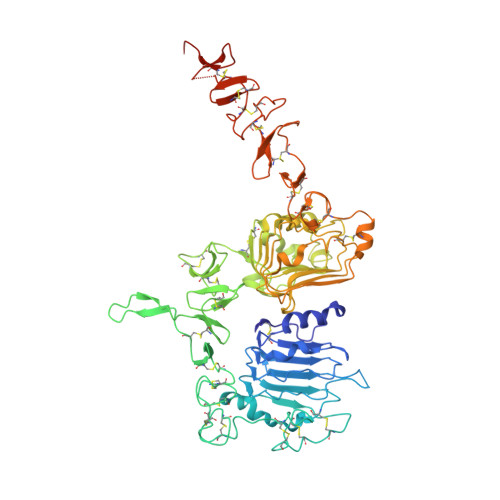
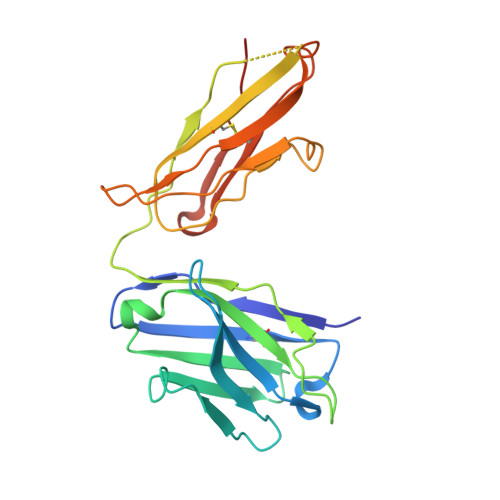
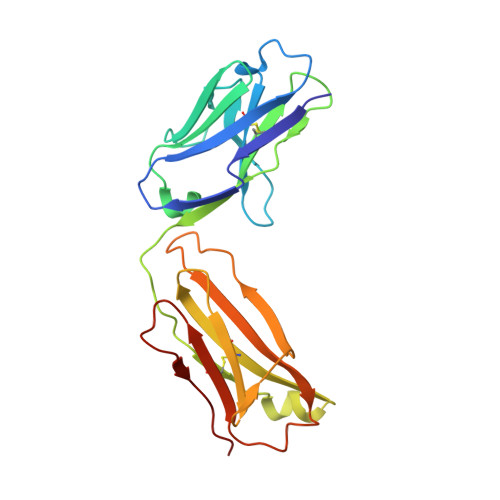
Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site
Bostrom, J., Yu, S.F., Kan, D., Appleton, B.A., Lee, C.V., Billeci, K., Man, W., Peale, F., Ross, S., Wiesmann, C., Fuh, G.(2009) Science 323: 1610-1614
- PubMed: 19299620
- DOI: https://doi.org/10.1126/science.1165480
- Primary Citation of Related Structures:
3BDY, 3BE1 - PubMed Abstract:
The interface between antibody and antigen is often depicted as a lock and key, suggesting that an antibody surface can accommodate only one antigen. Here, we describe an antibody with an antigen binding site that binds two distinct proteins with high affinity. We isolated a variant of Herceptin, a therapeutic monoclonal antibody that binds the human epidermal growth factor receptor 2 (HER2), on the basis of its ability to simultaneously interact with vascular endothelial growth factor (VEGF). Crystallographic and mutagenesis studies revealed that distinct amino acids of this antibody, called bH1, engage HER2 and VEGF energetically, but there is extensive overlap between the antibody surface areas contacting the two antigens. An affinity-improved version of bH1 inhibits both HER2- and VEGF-mediated cell proliferation in vitro and tumor progression in mouse models. Such "two-in-one" antibodies challenge the monoclonal antibody paradigm of one binding site, one antigen. They could also provide new opportunities for antibody-based therapy.
Organizational Affiliation:
Department of Protein Engineering, Genentech, 1 DNA Way, South San Francisco, CA 94080, USA.