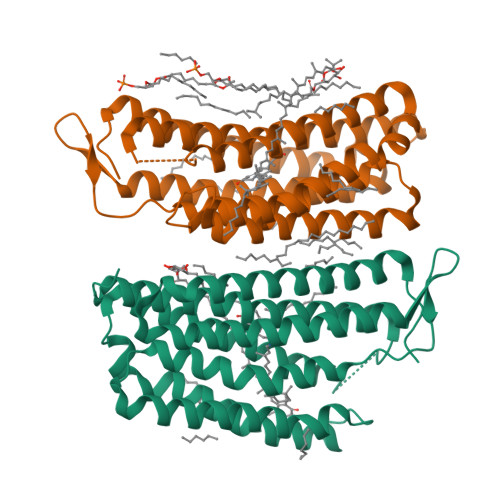
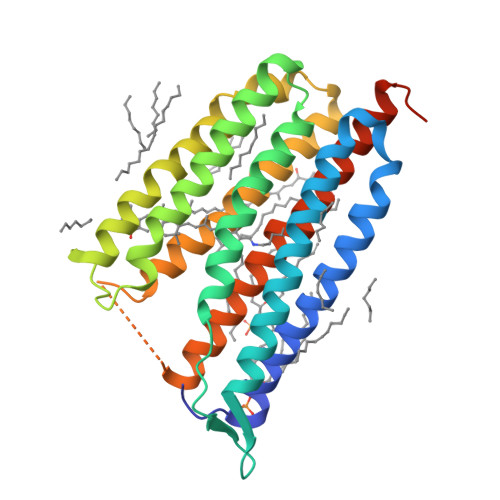
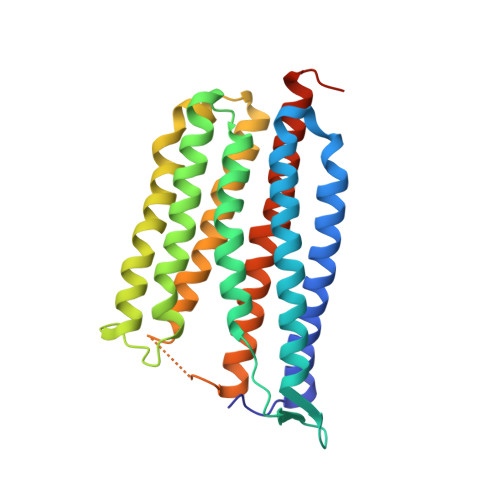
Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore.
Luecke, H., Schobert, B., Stagno, J., Imasheva, E.S., Wang, J.M., Balashov, S.P., Lanyi, J.K.(2008) Proc Natl Acad Sci U S A 105: 16561-16565
- PubMed: 18922772
- DOI: https://doi.org/10.1073/pnas.0807162105
- Primary Citation of Related Structures:
3DDL - PubMed Abstract:
Homologous to bacteriorhodopsin and even more to proteorhodopsin, xanthorhodopsin is a light-driven proton pump that, in addition to retinal, contains a noncovalently bound carotenoid with a function of a light-harvesting antenna. We determined the structure of this eubacterial membrane protein-carotenoid complex by X-ray diffraction, to 1.9-A resolution. Although it contains 7 transmembrane helices like bacteriorhodopsin and archaerhodopsin, the structure of xanthorhodopsin is considerably different from the 2 archaeal proteins. The crystallographic model for this rhodopsin introduces structural motifs for proton transfer during the reaction cycle, particularly for proton release, that are dramatically different from those in other retinal-based transmembrane pumps. Further, it contains a histidine-aspartate complex for regulating the pK(a) of the primary proton acceptor not present in archaeal pumps but apparently conserved in eubacterial pumps. In addition to aiding elucidation of a more general proton transfer mechanism for light-driven energy transducers, the structure defines also the geometry of the carotenoid and the retinal. The close approach of the 2 polyenes at their ring ends explains why the efficiency of the excited-state energy transfer is as high as approximately 45%, and the 46 degrees angle between them suggests that the chromophore location is a compromise between optimal capture of light of all polarization angles and excited-state energy transfer.
Organizational Affiliation:
Departments of Molecular Biology and Biochemistry, and Physiology and Biophysics, and Center for Biomembrane Systems, University of California, Irvine, CA 92697.