Structure-Function Relationships in Fungal Large-Subunit Catalases
Diaz, A., Valdes, V.-J., Rudino-Pinera, E., Horjales, E., Hansberg, W.(2008) J Mol Biology 386: 218-232
- PubMed: 19109972
- DOI: https://doi.org/10.1016/j.jmb.2008.12.019
- Primary Citation of Related Structures:
3EJ6 - PubMed Abstract:
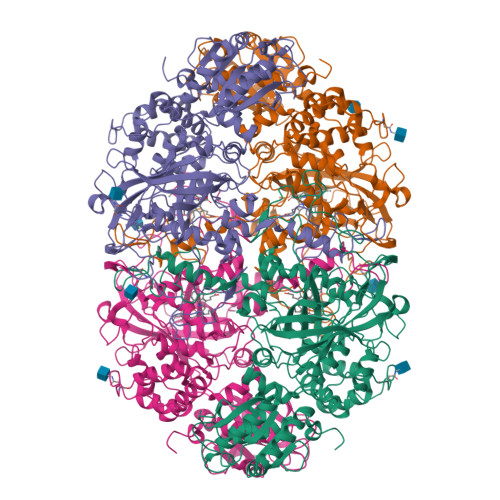
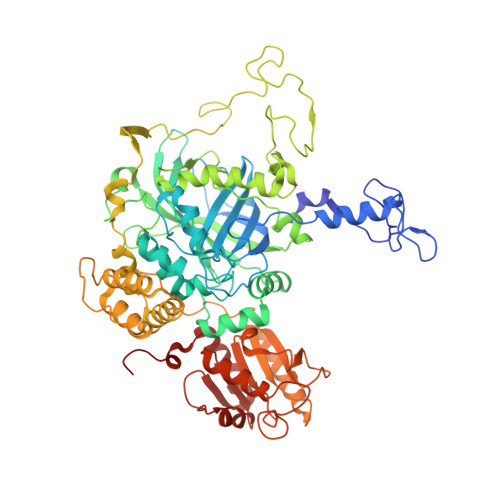
Neurospora crassa has two large-subunit catalases, CAT-1 and CAT-3. CAT-1 is associated with non-growing cells and accumulates particularly in asexual spores; CAT-3 is associated with growing cells and is induced under different stress conditions. It is our interest to elucidate the structure-function relationships in large-subunit catalases. Here we have determined the CAT-3 crystal structure and compared it with the previously determined CAT-1 structure. Similar to CAT-1, CAT-3 hydrogen peroxide (H(2)O(2)) saturation kinetics exhibited two components, consistent with the existence of two active sites: one saturated in the millimolar range and the other in the molar range. In the CAT-1 structure, we found three interesting features related to its unusual kinetics: (a) a constriction in the channel that conveys H(2)O(2) to the active site; (b) a covalent bond between the tyrosine, which forms the fifth coordination bound to the iron of the heme, and a vicinal cysteine; (c) oxidation of the pyrrole ring III to form a cis-hydroxyl group in C5 and a cis-gamma-spirolactone in C6. The site of heme oxidation marks the starts of the central channel that communicates to the central cavity and the shortest way products can exit the active site. CAT-3 has a similar constriction in its major channel, which could function as a gating system regulated by the H(2)O(2) concentration before the gate. CAT-3 functional tyrosine is not covalently bonded, but has instead the electron relay mechanism described for the human catalase to divert electrons from it. Pyrrole ring III in CAT-3 is not oxidized as it is in other large-subunit catalases whose structure has been determined. Different in CAT-3 from these enzymes is an occupied central cavity. Results presented here indicate that CAT-3 and CAT-1 enzymes represent a functional group of catalases with distinctive structural characteristics that determine similar kinetics.
Organizational Affiliation:
Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Circuito exterior s/n, Ciudad Universitaria, México, D. F. CP 04510, México.