An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
Kress, D., Brugel, D., Schall, I., Linder, D., Buckel, W., Essen, L.-O.(2009) J Biological Chem 284: 28401-28409
- PubMed: 19654317
- DOI: https://doi.org/10.1074/jbc.M109.037762
- Primary Citation of Related Structures:
3GF3, 3GF7, 3GLM, 3GMA - PubMed Abstract:
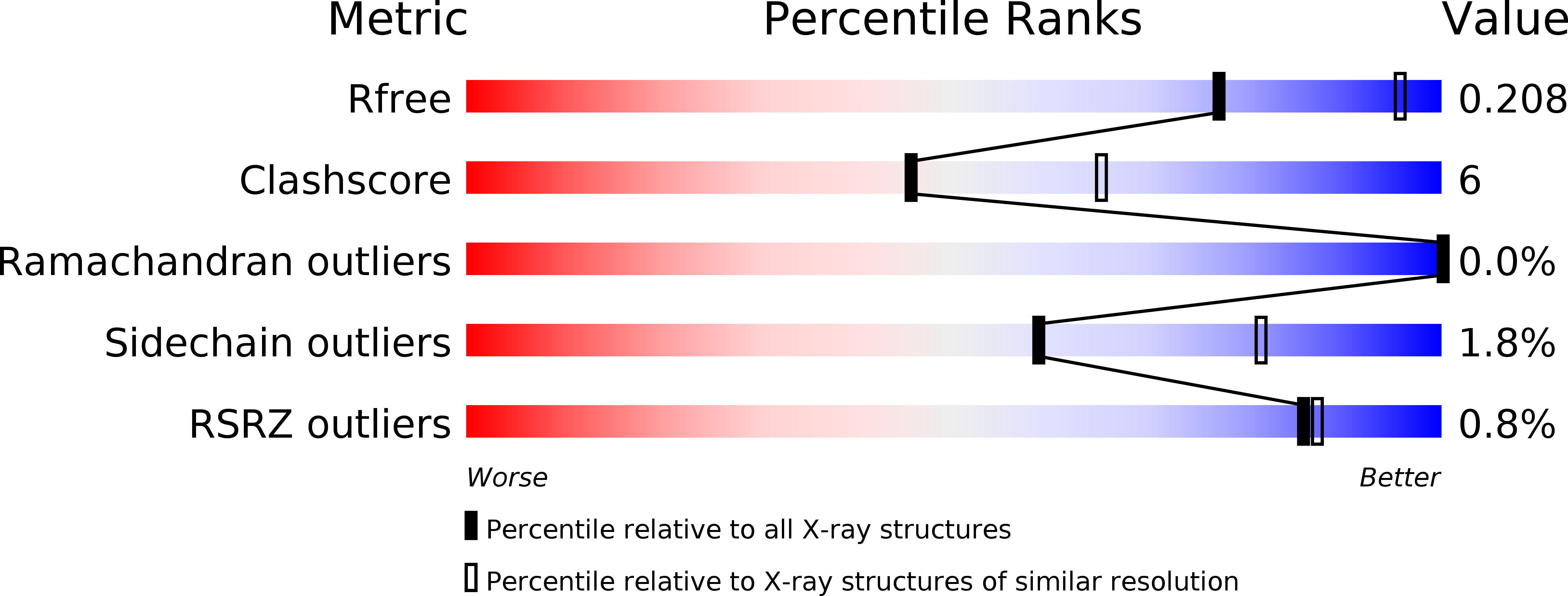
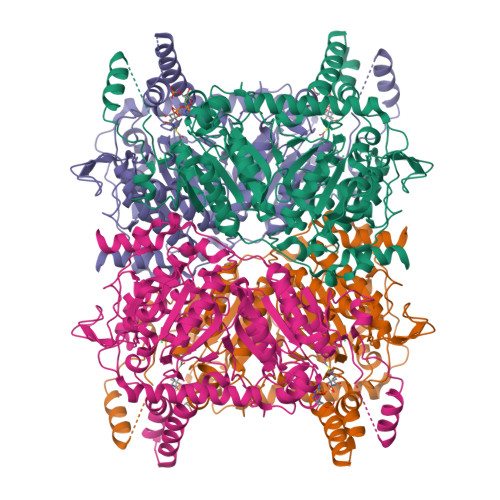
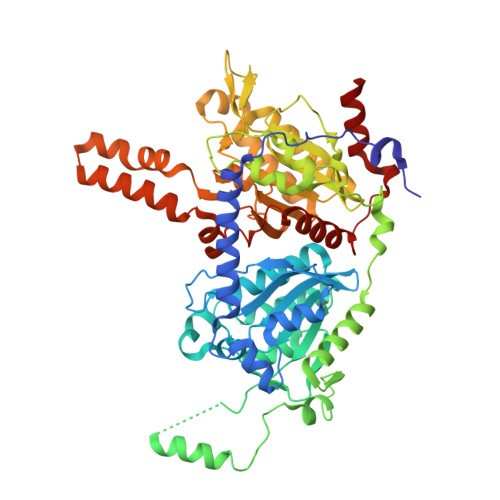
Glutaconyl-CoA decarboxylase (Gcd) couples the biotin-dependent decarboxylation of glutaconyl-CoA with the generation of an electrochemical Na(+) gradient. Sequencing of the genes encoding all subunits of the Clostridium symbiosum decarboxylase membrane complex revealed that it comprises two distinct biotin carrier subunits, GcdC(1) and GcdC(2), which differ in the length of a central alanine- and proline-rich linker domain. Co-crystallization of the decarboxylase subunit GcdA with the substrate glutaconyl-CoA, the product crotonyl-CoA, and the substrate analogue glutaryl-CoA, respectively, resulted in a high resolution model for substrate binding and catalysis revealing remarkable structural changes upon substrate binding. Unlike the GcdA structure from Acidaminococcus fermentans, these data suggest that in intact Gcd complexes, GcdA is associated as a tetramer crisscrossed by a network of solvent-filled tunnels.
Organizational Affiliation:
Biochemie, Fachbereich Chemie, Philipps-Universität Marburg, D-35032 Marburg, Germany.