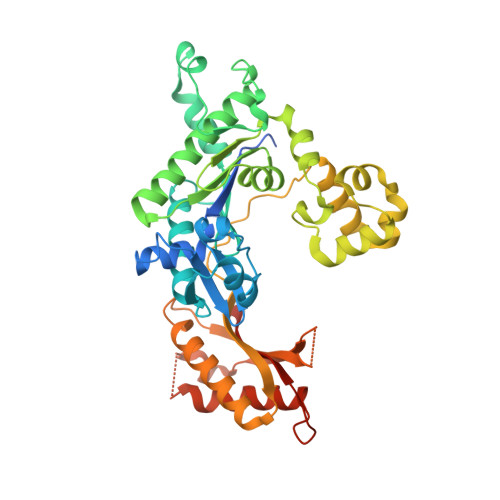
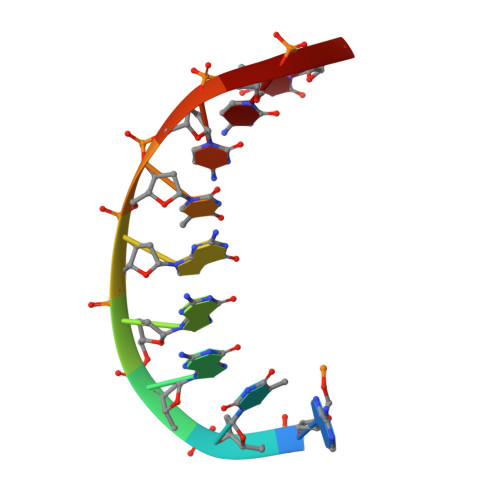
Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase iota
Kirouac, K.N., Ling, H.(2009) EMBO J 28: 1644-1654
- PubMed: 19440206
- DOI: https://doi.org/10.1038/emboj.2009.122
- Primary Citation of Related Structures:
3GV5, 3GV7, 3GV8 - PubMed Abstract:
Human DNA polymerase iota (pol iota) is a unique member of Y-family polymerases, which preferentially misincorporates nucleotides opposite thymines (T) and halts replication at T bases. The structural basis of the high error rates remains elusive. We present three crystal structures of pol complexed with DNA containing a thymine base, paired with correct or incorrect incoming nucleotides. A narrowed active site supports a pyrimidine to pyrimidine mismatch and excludes Watson-Crick base pairing by pol. The template thymine remains in an anti conformation irrespective of incoming nucleotides. Incoming ddATP adopts a syn conformation with reduced base stacking, whereas incorrect dGTP and dTTP maintain anti conformations with normal base stacking. Further stabilization of dGTP by H-bonding with Gln59 of the finger domain explains the preferential T to G mismatch. A template 'U-turn' is stabilized by pol and the methyl group of the thymine template, revealing the structural basis of T stalling. Our structural and domain-swapping experiments indicate that the finger domain is responsible for pol's high error rates on pyrimidines and determines the incorporation specificity.
Organizational Affiliation:
Department of Biochemistry, University of Western Ontario, London, Ontario, Canada.