Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Anastasiou, D., Yu, Y., Israelsen, W.J., Jiang, J.K., Boxer, M.B., Hong, B.S., Tempel, W., Dimov, S., Shen, M., Jha, A., Yang, H., Mattaini, K.R., Metallo, C.M., Fiske, B.P., Courtney, K.D., Malstrom, S., Khan, T.M., Kung, C., Skoumbourdis, A.P., Veith, H., Southall, N., Walsh, M.J., Brimacombe, K.R., Leister, W., Lunt, S.Y., Johnson, Z.R., Yen, K.E., Kunii, K., Davidson, S.M., Christofk, H.R., Austin, C.P., Inglese, J., Harris, M.H., Asara, J.M., Stephanopoulos, G., Salituro, F.G., Jin, S., Dang, L., Auld, D.S., Park, H.W., Cantley, L.C., Thomas, C.J., Vander Heiden, M.G.(2012) Nat Chem Biol 8: 839-847
- PubMed: 22922757
- DOI: https://doi.org/10.1038/nchembio.1060
- Primary Citation of Related Structures:
3ME3, 3U2Z - PubMed Abstract:
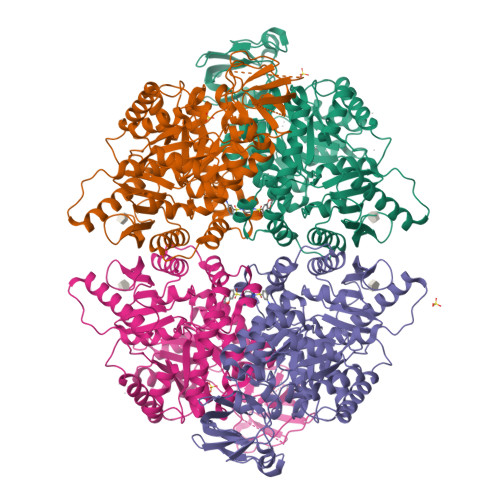
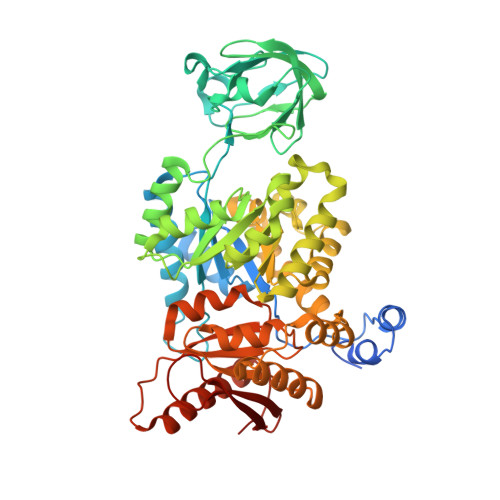
Cancer cells engage in a metabolic program to enhance biosynthesis and support cell proliferation. The regulatory properties of pyruvate kinase M2 (PKM2) influence altered glucose metabolism in cancer. The interaction of PKM2 with phosphotyrosine-containing proteins inhibits enzyme activity and increases the availability of glycolytic metabolites to support cell proliferation. This suggests that high pyruvate kinase activity may suppress tumor growth. We show that expression of PKM1, the pyruvate kinase isoform with high constitutive activity, or exposure to published small-molecule PKM2 activators inhibits the growth of xenograft tumors. Structural studies reveal that small-molecule activators bind PKM2 at the subunit interaction interface, a site that is distinct from that of the endogenous activator fructose-1,6-bisphosphate (FBP). However, unlike FBP, binding of activators to PKM2 promotes a constitutively active enzyme state that is resistant to inhibition by tyrosine-phosphorylated proteins. These data support the notion that small-molecule activation of PKM2 can interfere with anabolic metabolism.
Organizational Affiliation:
Department of Medicine, Division of Signal Transduction, Beth Israel Deaconess Medical Center, Boston, MA, USA.