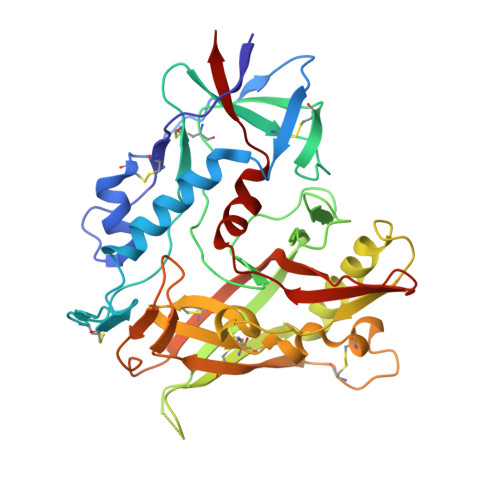
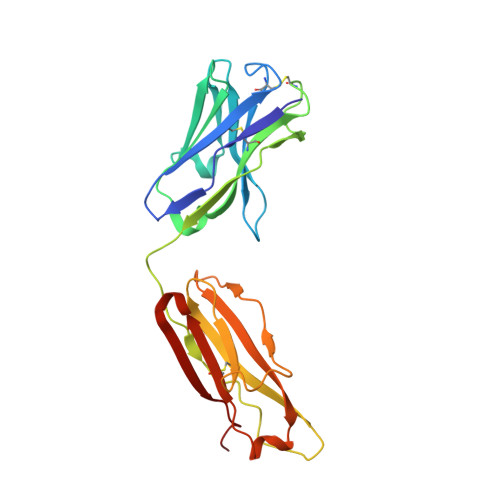
Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01.
Zhou, T., Georgiev, I., Wu, X., Yang, Z.Y., Dai, K., Finzi, A., Do Kwon, Y., Scheid, J.F., Shi, W., Xu, L., Yang, Y., Zhu, J., Nussenzweig, M.C., Sodroski, J., Shapiro, L., Nabel, G.J., Mascola, J.R., Kwong, P.D.(2010) Science 329: 811-817
- PubMed: 20616231
- DOI: https://doi.org/10.1126/science.1192819
- Primary Citation of Related Structures:
3NGB - PubMed Abstract:
During HIV-1 infection, antibodies are generated against the region of the viral gp120 envelope glycoprotein that binds CD4, the primary receptor for HIV-1. Among these antibodies, VRC01 achieves broad neutralization of diverse viral strains. We determined the crystal structure of VRC01 in complex with a human immunodeficiency virus HIV-1 gp120 core. VRC01 partially mimics CD4 interaction with gp120. A shift from the CD4-defined orientation, however, focuses VRC01 onto the vulnerable site of initial CD4 attachment, allowing it to overcome the glycan and conformational masking that diminishes the neutralization potency of most CD4-binding-site antibodies. To achieve this recognition, VRC01 contacts gp120 mainly through immunoglobulin V-gene regions substantially altered from their genomic precursors. Partial receptor mimicry and extensive affinity maturation thus facilitate neutralization of HIV-1 by natural human antibodies.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Bethesda, MD 20892, USA.