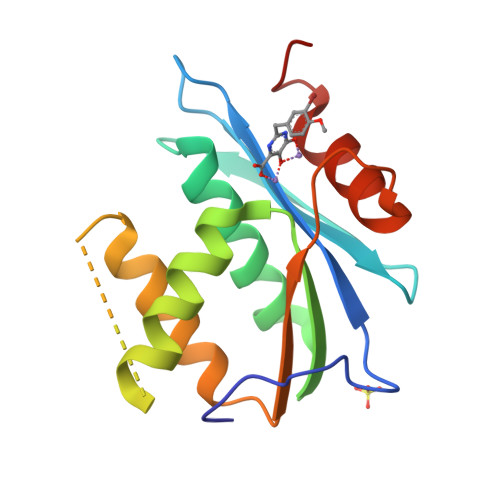
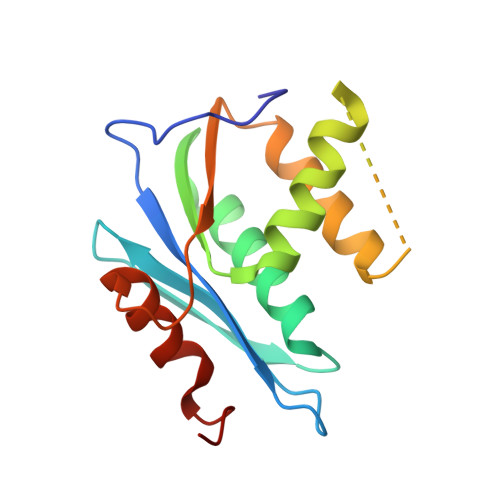
Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors.
Lansdon, E.B., Liu, Q., Leavitt, S.A., Balakrishnan, M., Perry, J.K., Lancaster-Moyer, C., Kutty, N., Liu, X., Squires, N.H., Watkins, W.J., Kirschberg, T.A.(2011) Antimicrob Agents Chemother 55: 2905-2915
- PubMed: 21464257
- DOI: https://doi.org/10.1128/AAC.01594-10
- Primary Citation of Related Structures:
3QIN, 3QIO, 3QIP - PubMed Abstract:
HIV-1 RNase H breaks down the intermediate RNA-DNA hybrids during reverse transcription, requiring two divalent metal ions for activity. Pyrimidinol carboxylic acid and N-hydroxy quinazolinedione inhibitors were designed to coordinate the two metal ions in the active site of RNase H. High-resolution (1.4 Å to 2.1 Å) crystal structures were determined with the isolated RNase H domain and reverse transcriptase (RT), which permit accurate assessment of the metal and water environment at the active site. The geometry of the metal coordination suggests that the inhibitors mimic a substrate state prior to phosphodiester catalysis. Surface plasmon resonance studies confirm metal-dependent binding to RNase H and demonstrate that the inhibitors do not bind at the polymerase active site of RT. Additional evaluation of the RNase H site reveals an open protein surface with few additional interactions to optimize active-site inhibitors.
Organizational Affiliation:
Gilead Sciences, Inc., 333 Lakeside Dr., Foster City, CA 94404, USA. eric.lansdon@gilead.com