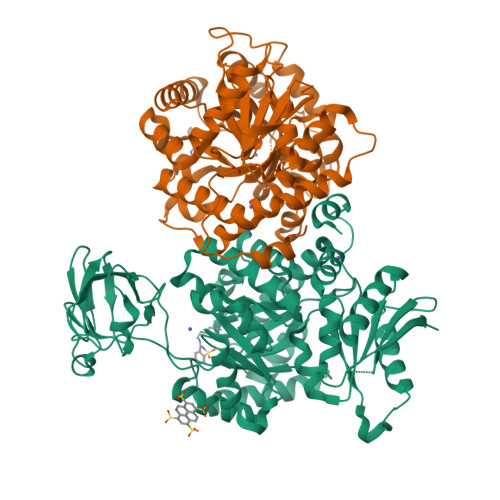
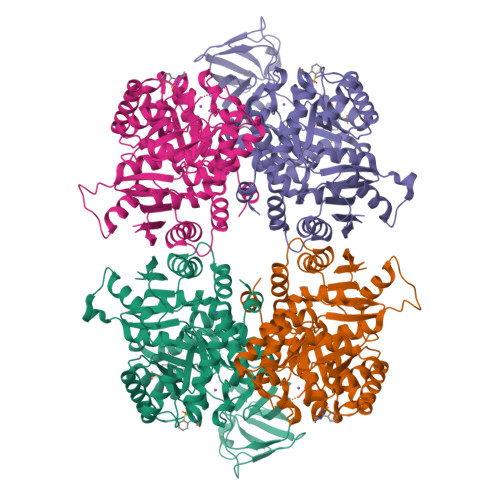
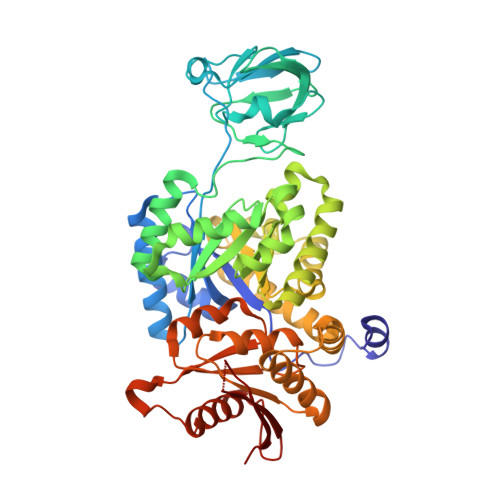
A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
Morgan, H.P., Walsh, M., Blackburn, E.A., Wear, M.A., Boxer, M., Shen, M., McNae, I.W., Michels, P.A.M., Auld, D.S., Fothergill-Gilmore, L.A., Walkinshaw, M.D.To be published.