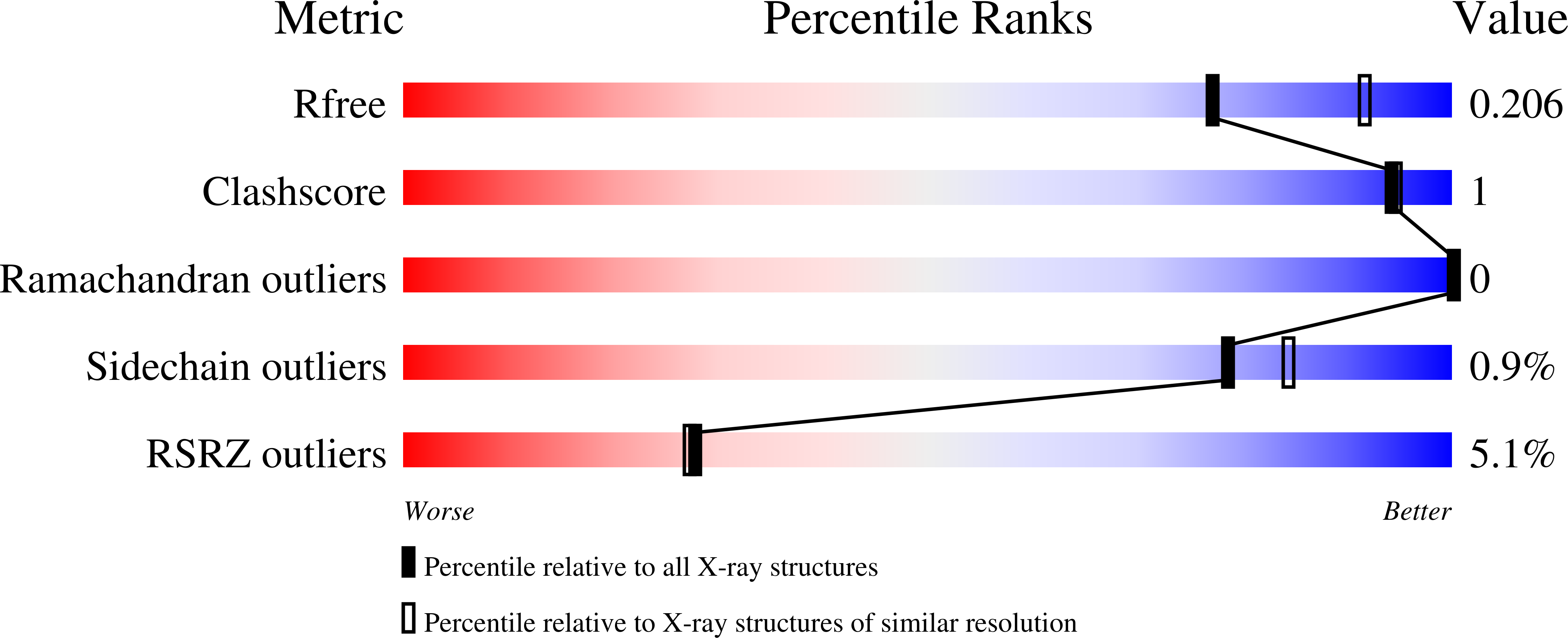
A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
Akey, D.L., Li, S., Konwerski, J.R., Confer, L.A., Bernard, S.M., Anzai, Y., Kato, F., Sherman, D.H., Smith, J.L.(2011) J Mol Biol 413: 438-450
- PubMed: 21884704
- DOI: https://doi.org/10.1016/j.jmb.2011.08.040
- Primary Citation of Related Structures:
3SSM, 3SSN, 3SSO - PubMed Abstract:
O-linked methylation of sugar substituents is a common modification in the biosynthesis of many natural products and is catalyzed by multiple families of S-adenosyl-L-methionine (SAM or AdoMet)-dependent methyltransferases (MTs). Mycinamicins, potent antibiotics from Micromonospora griseorubida, can be methylated at two positions on a 6-deoxyallose substituent. The first methylation is catalyzed by MycE, a SAM- and metal-dependent MT. Crystal structures were determined for MycE bound to the product S-adenosyl-L-homocysteine (AdoHcy) and magnesium, both with and without the natural substrate mycinamicin VI. This represents the first structure of a natural product sugar MT in complex with its natural substrate. MycE is a tetramer of a two-domain polypeptide, comprising a C-terminal catalytic MT domain and an N-terminal auxiliary domain, which is important for quaternary assembly and for substrate binding. The symmetric MycE tetramer has a novel MT organization in which each of the four active sites is formed at the junction of three monomers within the tetramer. The active-site structure supports a mechanism in which a conserved histidine acts as a general base, and the metal ion helps to position the methyl acceptor and to stabilize a hydroxylate intermediate. A conserved tyrosine is suggested to support activity through interactions with the transferred methyl group from the SAM methyl donor. The structure of the free enzyme reveals a dramatic order-disorder transition in the active site relative to the S-adenosyl-L-homocysteine complexes, suggesting a mechanism for product/substrate exchange through concerted movement of five loops and the polypeptide C-terminus.
Organizational Affiliation:
Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109, USA.