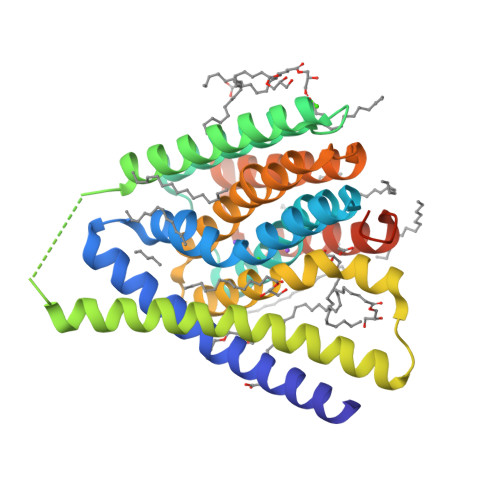
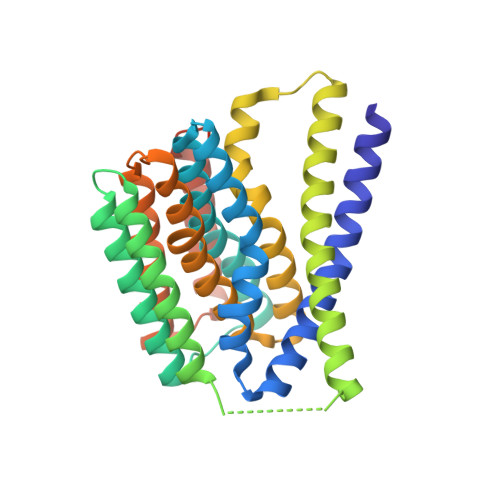
Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Liao, J., Li, H., Zeng, W., Sauer, D.B., Belmares, R., Jiang, Y.(2012) Science 335: 686-690
- PubMed: 22323814
- DOI: https://doi.org/10.1126/science.1215759
- Primary Citation of Related Structures:
3V5S, 3V5U - PubMed Abstract:
Sodium/calcium (Na(+)/Ca(2+)) exchangers (NCX) are membrane transporters that play an essential role in maintaining the homeostasis of cytosolic Ca(2+) for cell signaling. We demonstrated the Na(+)/Ca(2+)-exchange function of an NCX from Methanococcus jannaschii (NCX_Mj) and report its 1.9 angstrom crystal structure in an outward-facing conformation. Containing 10 transmembrane helices, the two halves of NCX_Mj share a similar structure with opposite orientation. Four ion-binding sites cluster at the center of the protein: one specific for Ca(2+) and three that likely bind Na(+). Two passageways allow for Na(+) and Ca(2+) access to the central ion-binding sites from the extracellular side. Based on the symmetry of NCX_Mj and its ability to catalyze bidirectional ion-exchange reactions, we propose a structure model for the inward-facing NCX_Mj.
Organizational Affiliation:
Department of Physiology, University of Texas Southwestern Medical Center, Dallas, TX 75390-9040, USA.