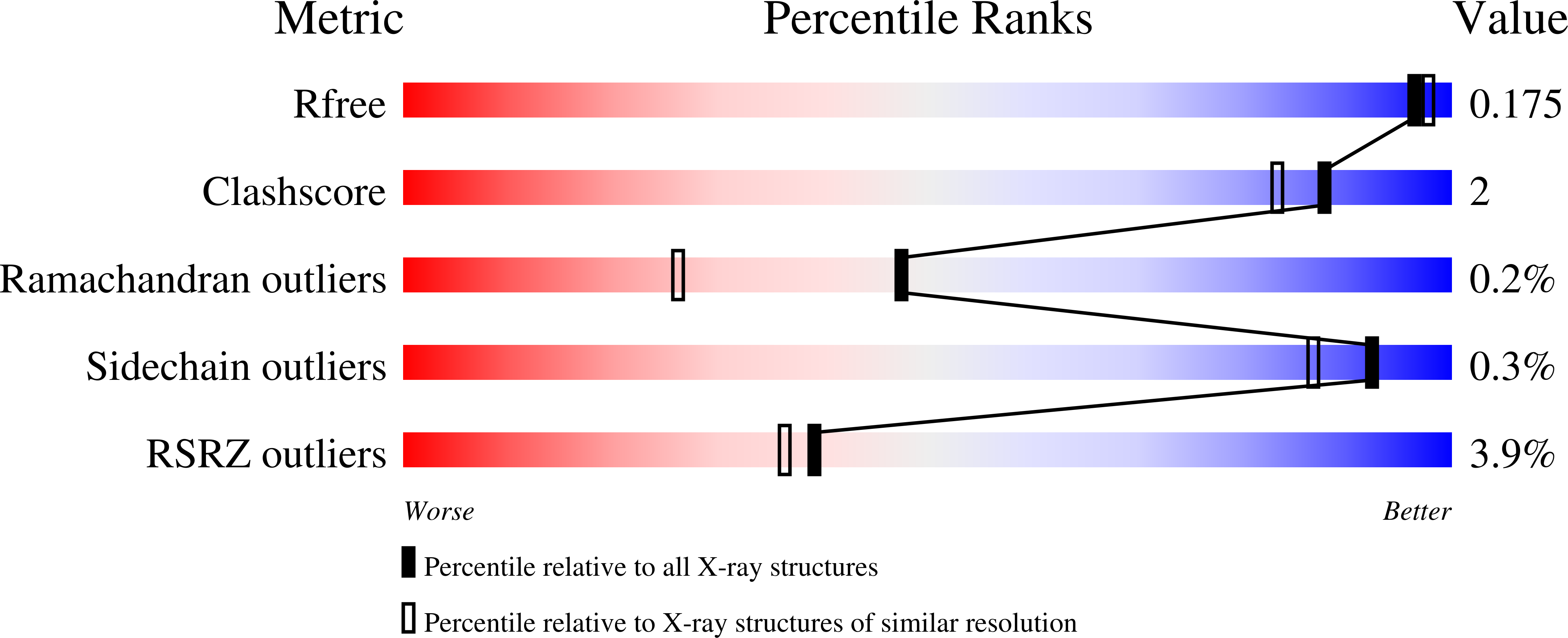
Structure and Catalytic Mechanism of 3-Ketosteroid-{Delta}4-(5Alpha)-Dehydrogenase from Rhodococcus Jostii Rha1 Genome.
Van Oosterwijk, N., Knol, J., Dijkhuizen, L., Van Der Geize, R., Dijkstra, B.W.(2012) J Biol Chem 287: 30975
- PubMed: 22833669
- DOI: https://doi.org/10.1074/jbc.M112.374306
- Primary Citation of Related Structures:
4AT0, 4AT2 - PubMed Abstract:
3-Ketosteroid Δ4-(5α)-dehydrogenases (Δ4-(5α)-KSTDs) are enzymes that introduce a double bond between the C4 and C5 atoms of 3-keto-(5α)-steroids. Here we show that the ro05698 gene from Rhodococcus jostii RHA1 codes for a flavoprotein with Δ4-(5α)-KSTD activity. The 1.6 Å resolution crystal structure of the enzyme revealed three conserved residues (Tyr-319, Tyr-466, and Ser-468) in a pocket near the isoalloxazine ring system of the FAD co-factor. Site-directed mutagenesis of these residues confirmed that they are absolutely essential for catalytic activity. A crystal structure with bound product 4-androstene-3,17-dione showed that Ser-468 is in a position in which it can serve as the base abstracting the 4β-proton from the C4 atom of the substrate. Ser-468 is assisted by Tyr-319, which possibly is involved in shuttling the proton to the solvent. Tyr-466 is at hydrogen bonding distance to the C3 oxygen atom of the substrate and can stabilize the keto-enol intermediate occurring during the reaction. Finally, the FAD N5 atom is in a position to be able to abstract the 5α-hydrogen of the substrate as a hydride ion. These features fully explain the reaction catalyzed by Δ4-(5α)-KSTDs.
Organizational Affiliation:
Laboratory of Biophysical Chemistry, University of Groningen, 9747 AG Groningen, The Netherlands.