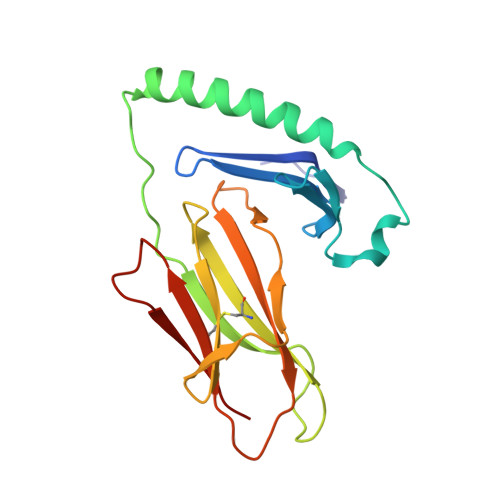
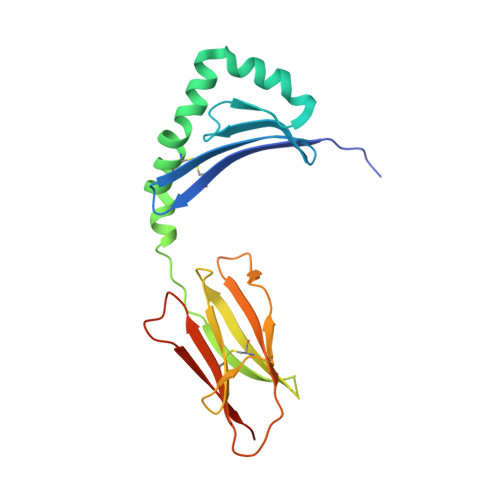
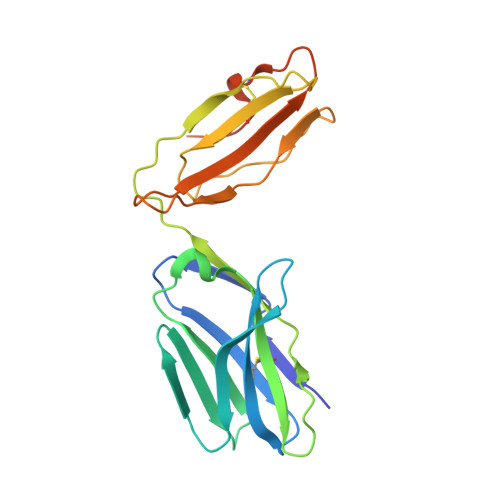
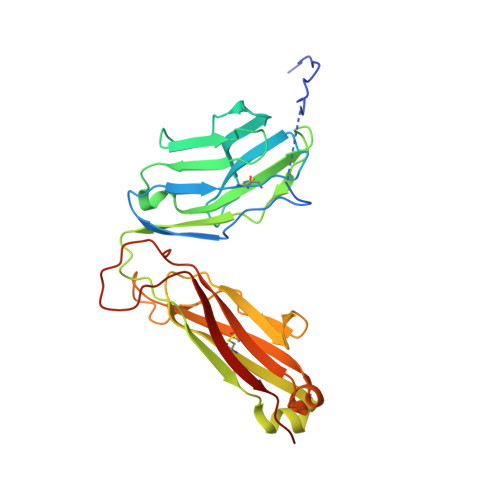
Crossreactivity of a human autoimmune TCR is dominated by a single TCR loop.
Sethi, D.K., Gordo, S., Schubert, D.A., Wucherpfennig, K.W.(2013) Nat Commun 4: 2623-2623
- PubMed: 24136005
- DOI: https://doi.org/10.1038/ncomms3623
- Primary Citation of Related Structures:
4GRL, 4MAY - PubMed Abstract:
Self-reactive CD4 T cells are thought to have a central role in the pathogenesis of many chronic inflammatory human diseases. Microbial peptides can activate self-reactive T cells, but the structural basis for such crossreactivity is not well understood. The Hy.1B11 T cell receptor (TCR) originates from a patient with multiple sclerosis and recognizes the self-antigen myelin basic protein. Here we report the structural mechanism of TCR crossreactivity with two distinct peptides from human pathogens. The structures show that a single TCR residue (CDR3α F95) makes the majority of contacts with the self-peptide and both microbial peptides (66.7-80.6%) due to a highly tilted TCR-binding topology on the peptide-MHC surface. Further, a neighbouring residue located on the same TCR loop (CDR3α E98) forms an energetically critical interaction with the MHC molecule. These data show how binding by a self-reactive TCR favors crossreactivity between self and microbial antigens.
Organizational Affiliation:
Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts 02115, USA.