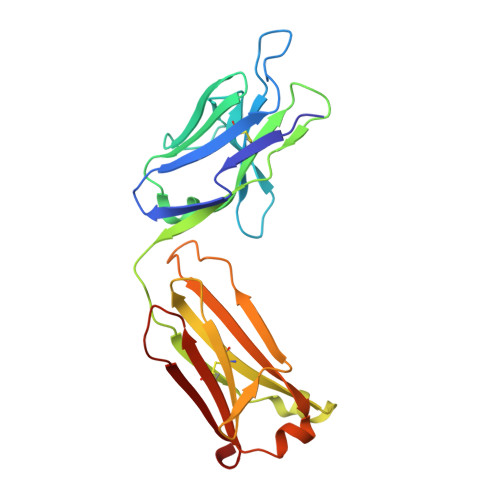
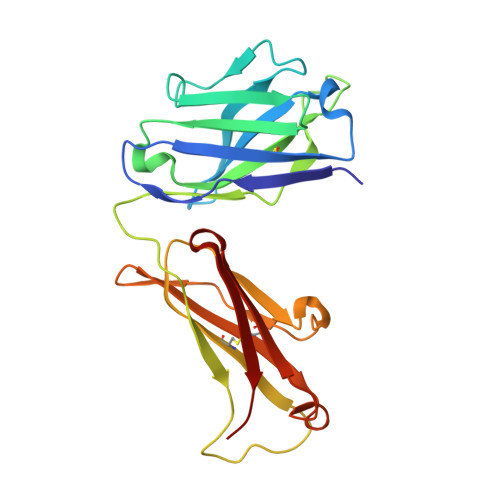
Global structures of IgG isotypes expressing identical variable regions.
Eryilmaz, E., Janda, A., Kim, J., Cordero, R.J., Cowburn, D., Casadevall, A.(2013) Mol Immunol 56: 588-598
- PubMed: 23911417
- DOI: https://doi.org/10.1016/j.molimm.2013.06.006
- Primary Citation of Related Structures:
4HDI - PubMed Abstract:
Until relatively recently the immunoglobulin molecule was viewed as composed of two independent domains comprised of the variable (V) and constant (C) regions. However, recent work has established that the C region mediates allosteric changes in the V region that can influence specificity and affinity. To further explore cross-domain interrelationship in murine IgG structure we carried out solution small angle X-ray scattering (SAXS) measurements for four V region identical IgG isotypes. SAXS analysis revealed elongated Y-shaped structures in solution with significantly different, isotype-dependent domain orientations. To further explore local C region effects on the V region, the IgG₃ Fab crystal structure from the same family was determined to 2.45 Å resolution. The IgG₃ Fab crystal structure differs from a closely related previously solved IgG1 Fab revealing significant structural differences, which may account for isotype-related specificity differences in V region identical Abs. Among the four murine isotypes, IgG₃ was the most different in solution with regards to overall structure as well as aggregate formation in solution suggesting that the greater apparent affinity of this isotype resulted from polyvalent complexes with enhanced avidity. Our results provide additional evidence that Ig V and C domains influence each other structurally and suggest that V region structure can have significant effects on overall Ig structure.
Organizational Affiliation:
Department of Biochemistry, The Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA.