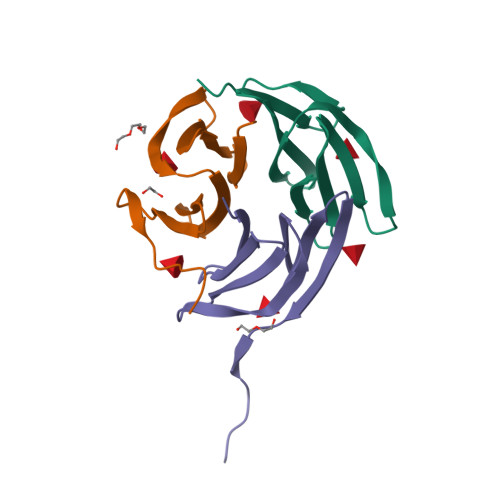
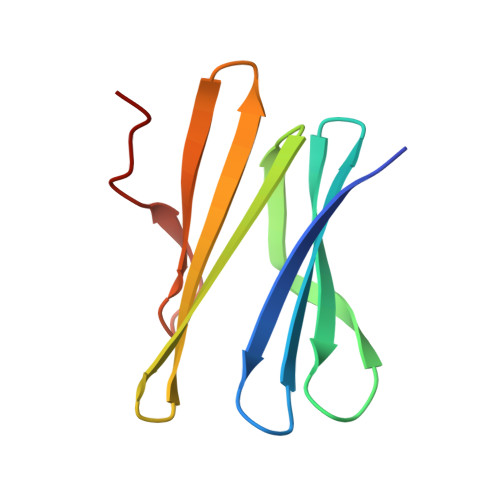
Reduction of lectin valency drastically changes glycolipid dynamics in membranes but not surface avidity
Arnaud, J., Claudinon, J., Trondle, K., Trovaslet, M., Larson, G., Thomas, A., Varrot, A., Romer, W., Imberty, A., Audfray, A.(2013) ACS Chem Biol 8: 1918-1924
- PubMed: 23855446
- DOI: https://doi.org/10.1021/cb400254b
- Primary Citation of Related Structures:
3ZI8, 4I6S - PubMed Abstract:
Multivalency is proposed to play a role in the strong avidity of lectins for glycosylated cell surfaces and also in their ability to affect membrane dynamics by clustering glycosphingolipids. Lectins with modified valency were designed from the β-propeller fold of Ralstonia solanacearum lectin (RSL) that presents six fucose binding sites. After identification of key amino acids by molecular dynamics calculations, two mutants with reduced valency were produced. Isothermal titration calorimetry confirmed the loss of three high affinity binding sites for both mutants. Crystal structures indicated that residual low affinity binding occurred in W76A but not in R17A. The trivalent R17A mutant presented unchanged avidity toward fucosylated surfaces, when compared to hexavalent RSL. However, R17A is not able anymore to induce formation of membrane invaginations on giant unilamellar vesicules, indicating the crucial role of number of binding sites for clustering of glycolipids. In the human lung epithelial cell line H1299, wt-RSL is internalized within seconds whereas the kinetics of R17A uptake is largely delayed. Neolectins with tailored valency are promising tools to study membrane dynamics.
Organizational Affiliation:
CERMAV-CNRS (affiliated to Grenoble Université and ICMG), BP53, 38041 Grenoble, France.