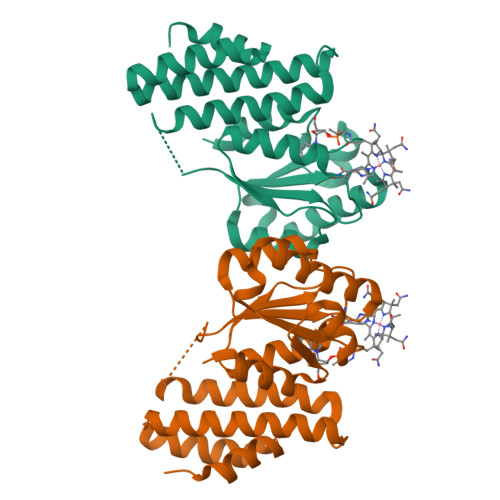
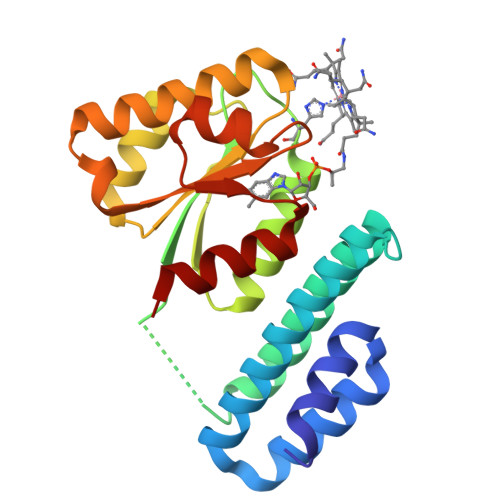
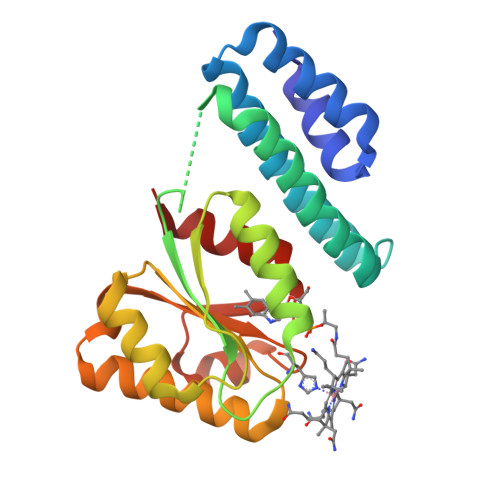
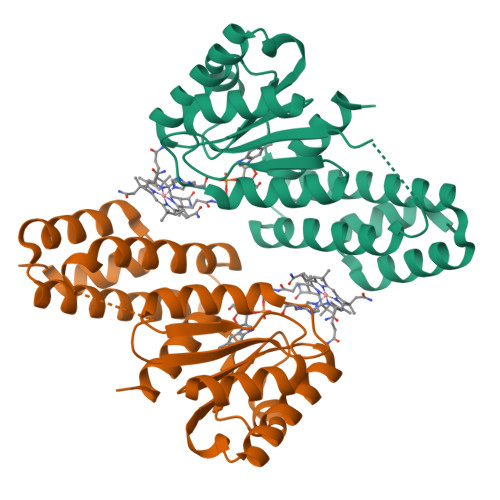
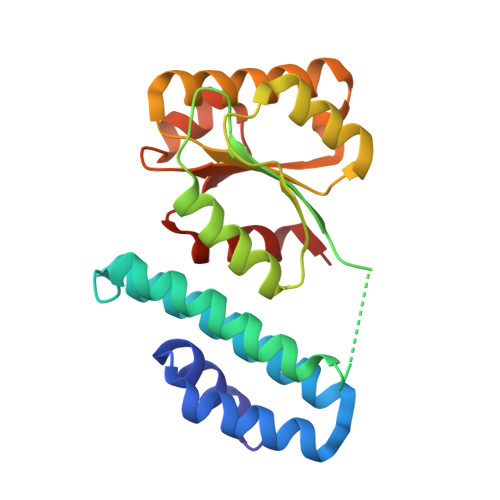
Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Sjuts, H., Dunstan, M.S., Fisher, K., Leys, D.(2013) Acta Crystallogr D Biol Crystallogr 69: 1609-1616
- PubMed: 23897483
- DOI: https://doi.org/10.1107/S0907444913011323
- Primary Citation of Related Structures:
4JGI - PubMed Abstract:
This study describes the identification and the structural and spectroscopic analysis of a cobalamin-binding protein (termed CobDH) implicated in O-demethylation by the organohalide-respiring bacterium Desulfitobacterium hafniense DCB-2. The 1.5 Å resolution crystal structure of CobDH is presented in the cobalamin-bound state and reveals that the protein is composed of an N-terminal helix-bundle domain and a C-terminal Rossmann-fold domain, with the cobalamin coordinated in the base-off/His-on conformation similar to other cobalamin-binding domains that catalyse methyl-transfer reactions. EPR spectroscopy of CobDH confirms cobalamin binding and reveals the presence of a cob(III)alamin superoxide, indicating binding of oxygen to the fully oxidized cofactor. These data provide the first structural insights into the methyltransferase reactions that occur during O-demethylation by D. hafniense.
Organizational Affiliation:
Manchester Institute of Biotechnology, Faculty of Life Sciences, University of Manchester, 131 Princess Street, Manchester M1 7DN, England.