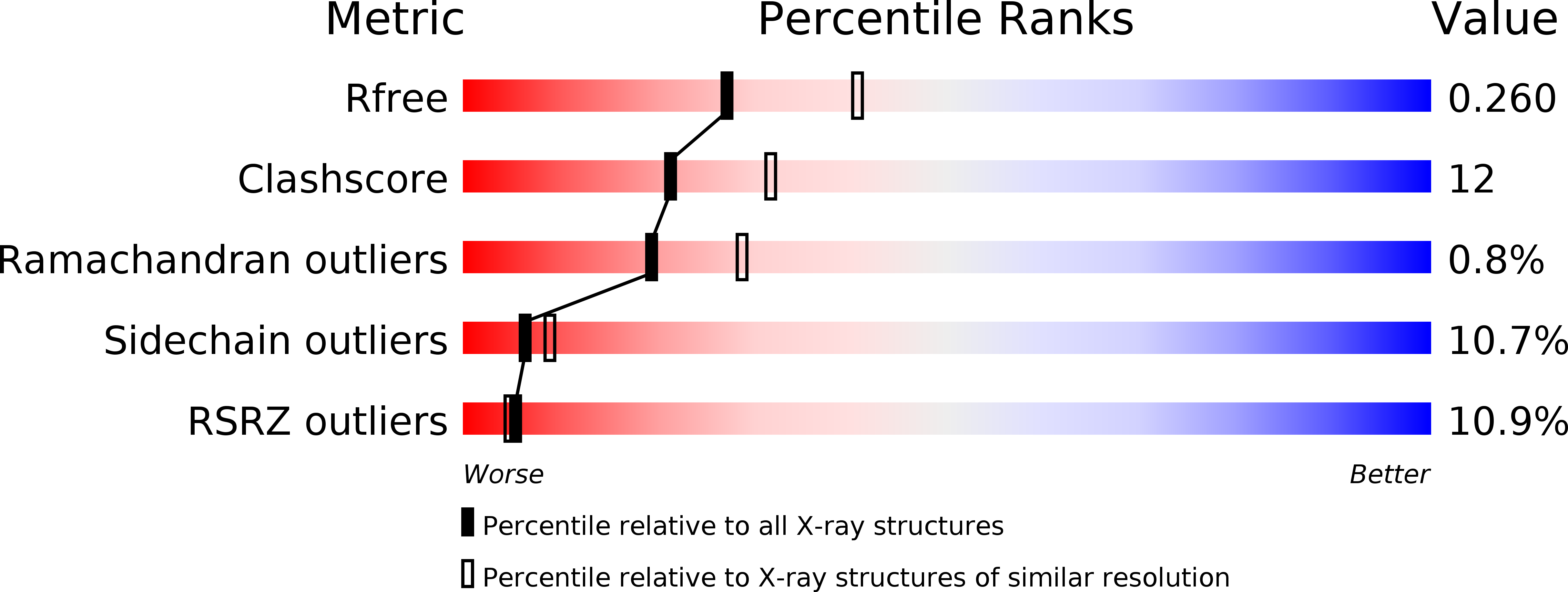
Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut).
Lu, X., Shi, Y., Zhang, W., Zhang, Y., Qi, J., Gao, G.F.(2013) Protein Cell 4: 502-511
- PubMed: 23794001
- DOI: https://doi.org/10.1007/s13238-013-3906-z
- Primary Citation of Related Structures:
4KDM, 4KDN, 4KDO, 4KDQ - PubMed Abstract:
Avian influenza A virus continues to pose a global threat with occasional H5N1 human infections, which is emphasized by a recent severe human infection caused by avian-origin H7N9 in China. Luckily these viruses do not transmit efficiently in human populations. With a few amino acid substitutions of the hemagglutinin H5 protein in the laboratory, two H5 mutants have been shown to obtain an air-borne transmission in a mammalian ferret model. Here in this study one of the mutant H5 proteins developed by Kawaoka's group (VN1203mut) was expressed in a baculovirus system and its receptor-binding properties were assessed. We herein show that the VN1203mut had a dramatically reduced binding affinity for the avian α2,3-linkage receptor compared to wild type but showed no detectable increase in affinity for the human α2,6-linkage receptor, using Surface Plasmon Resonance techonology. Further, the crystal structures of the VN1203mut and its complexes with either human or avian receptors demonstrate that the VN1203mut binds the human receptor in the same binding manner (cis conformation) as seen for the HAs of previously reported 1957 and 1968 pandemic influenza viruses. Our receptor binding and crystallographic data shown here further confirm that the ability to bind the avian receptor has to decrease for a higher human receptor binding affinity. As the Q226L substitution is shown important for obtaining human receptor binding, we suspect that the newly emerged H7N9 binds human receptor as H7 has a Q226L substitution.
Organizational Affiliation:
College of Veterinary Medicine, China Agricultural University, Beijing 100193, China.