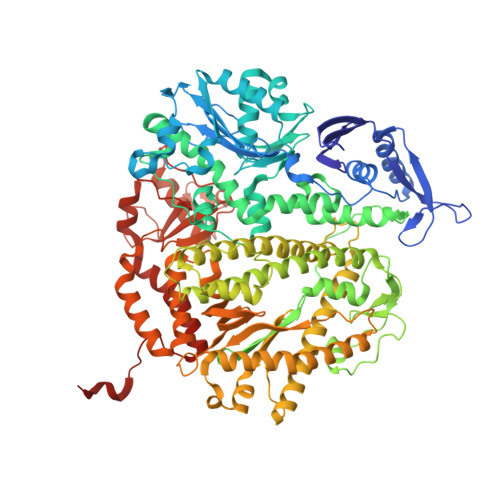
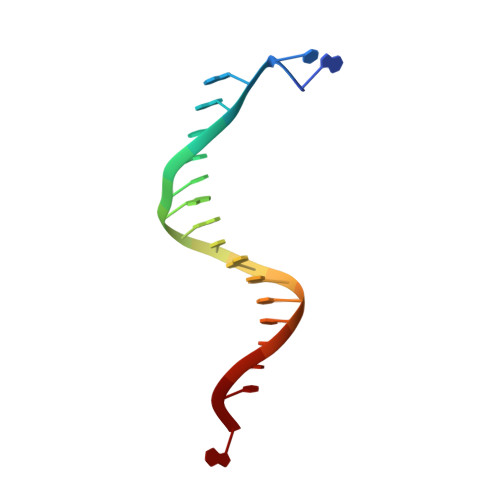
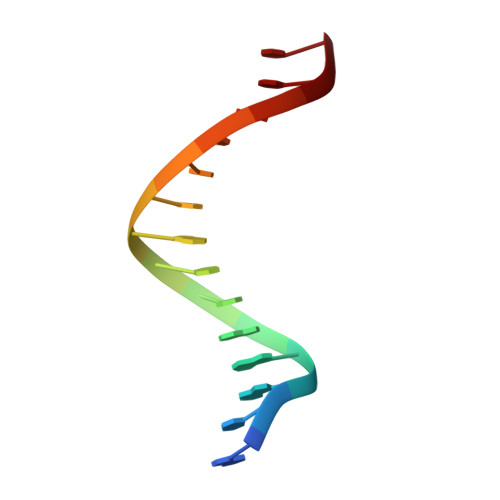
Structure-function analysis of ribonucleotide bypass by B family DNA replicases.
Clausen, A.R., Murray, M.S., Passer, A.R., Pedersen, L.C., Kunkel, T.A.(2013) Proc Natl Acad Sci U S A 110: 16802-16807
- PubMed: 24082122
- DOI: https://doi.org/10.1073/pnas.1309119110
- Primary Citation of Related Structures:
4KHQ, 4KHS, 4KHU, 4KHW, 4KHY, 4KI4, 4KI6 - PubMed Abstract:
Ribonucleotides are frequently incorporated into DNA during replication, they are normally removed, and failure to remove them results in replication stress. This stress correlates with DNA polymerase (Pol) stalling during bypass of ribonucleotides in DNA templates. Here we demonstrate that stalling by yeast replicative Pols δ and ε increases as the number of consecutive template ribonucleotides increases from one to four. The homologous bacteriophage RB69 Pol also stalls during ribonucleotide bypass, with a pattern most similar to that of Pol ε. Crystal structures of an exonuclease-deficient variant of RB69 Pol corresponding to multiple steps in single ribonucleotide bypass reveal that increased stalling is associated with displacement of Tyr391 and an unpreferred C2'-endo conformation for the ribose. Even less efficient bypass of two consecutive ribonucleotides in DNA correlates with similar movements of Tyr391 and displacement of one of the ribonucleotides along with the primer-strand DNA backbone. These structure-function studies have implications for cellular signaling by ribonucleotides, and they may be relevant to replication stress in cells defective in ribonucleotide excision repair, including humans suffering from autoimmune disease associated with RNase H2 defects.
Organizational Affiliation:
Laboratory of Structural Biology and Laboratory of Molecular Genetics, National Institute of Environmental Health Sciences, National Institute of Health, Department of Health and Human Services, Research Triangle Park, NC 27709.