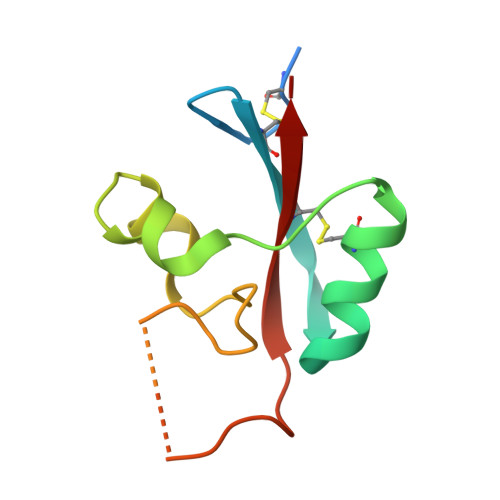
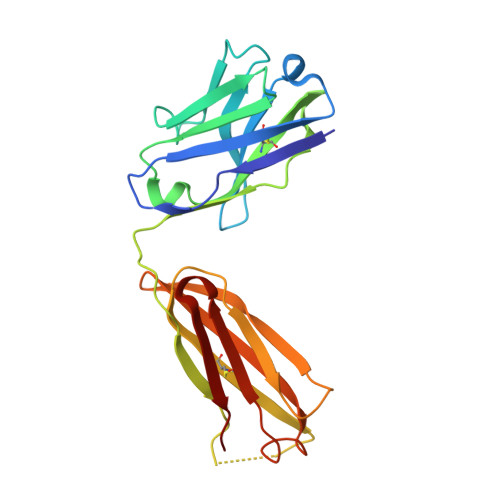
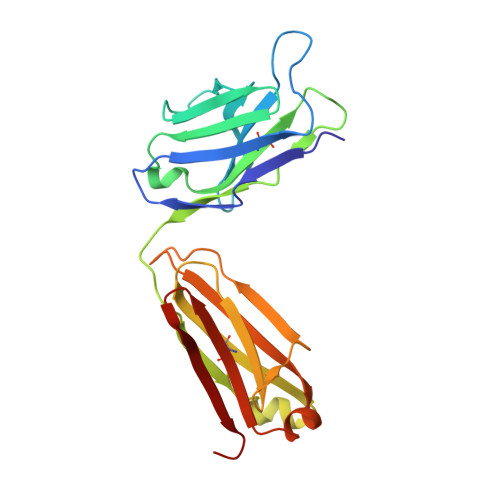
Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer.
Matho, M.H., Schlossman, A., Meng, X., Benhnia, M.R., Kaever, T., Buller, M., Doronin, K., Parker, S., Peters, B., Crotty, S., Xiang, Y., Zajonc, D.M.(2015) PLoS Pathog 11: e1005148-e1005148
- PubMed: 26325270
- DOI: https://doi.org/10.1371/journal.ppat.1005148
- Primary Citation of Related Structures:
4LQF, 4LU5, 4M1G - PubMed Abstract:
Vaccinia virus A33 is an extracellular enveloped virus (EEV)-specific type II membrane glycoprotein that is essential for efficient EEV formation and long-range viral spread within the host. A33 is a target for neutralizing antibody responses against EEV. In this study, we produced seven murine anti-A33 monoclonal antibodies (MAbs) by immunizing mice with live VACV, followed by boosting with the soluble A33 homodimeric ectodomain. Five A33 specific MAbs were capable of neutralizing EEV in the presence of complement. All MAbs bind to conformational epitopes on A33 but not to linear peptides. To identify the epitopes, we have adetermined the crystal structures of three representative neutralizing MAbs in complex with A33. We have further determined the binding kinetics for each of the three antibodies to wild-type A33, as well as to engineered A33 that contained single alanine substitutions within the epitopes of the three crystallized antibodies. While the Fab of both MAbs A2C7 and A20G2 binds to a single A33 subunit, the Fab from MAb A27D7 binds to both A33 subunits simultaneously. A27D7 binding is resistant to single alanine substitutions within the A33 epitope. A27D7 also demonstrated high-affinity binding with recombinant A33 protein that mimics other orthopoxvirus strains in the A27D7 epitope, such as ectromelia, monkeypox, and cowpox virus, suggesting that A27D7 is a potent cross-neutralizer. Finally, we confirmed that A27D7 protects mice against a lethal challenge with ectromelia virus.
Organizational Affiliation:
Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, California, United States of America.