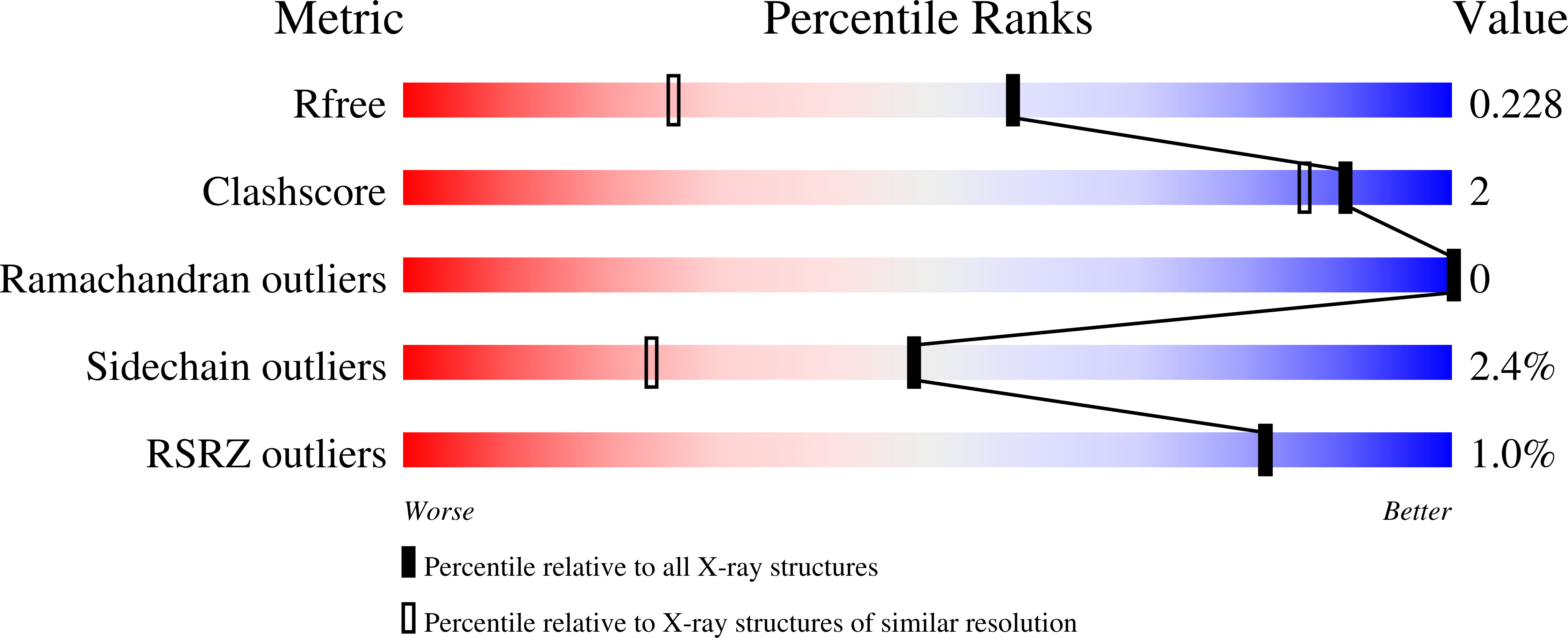
Production of a novel N-monomethylated dideoxysugar.
Thoden, J.B., Holden, H.M.(2014) Biochemistry 53: 1105-1107
- PubMed: 24512254
- DOI: https://doi.org/10.1021/bi500098a
- Primary Citation of Related Structures:
4OQD, 4OQE - PubMed Abstract:
The importance of unusual deoxysugars in biology has become increasingly apparent over the past decade. Some, for example, play key roles in the physiological activities of the natural products to which they are attached. Here we describe a study of TylM1, a dimethyltransferase from Streptomyces fradiae involved in the production of dTDP-mycaminose. From this investigation, the manner in which the enzyme binds its dimethylated product has been revealed. More significantly, by providing the enzyme with an alternative substrate, it was possible to produce a monomethylated product not observed in nature. This has important ramifications for the production of unique carbohydrates that may prove useful in drug design.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin , Madison, Wisconsin 53706, United States.