Transthyretin complexes with curcumin and bromo-estradiol: evaluation of solubilizing multicomponent mixtures.
Ciccone, L., Tepshi, L., Nencetti, S., Stura, E.A.(2014) N Biotechnol 32: 54-64
- PubMed: 25224922
- DOI: https://doi.org/10.1016/j.nbt.2014.09.002
- Primary Citation of Related Structures:
4PM1, 4PME, 4PMF - PubMed Abstract:
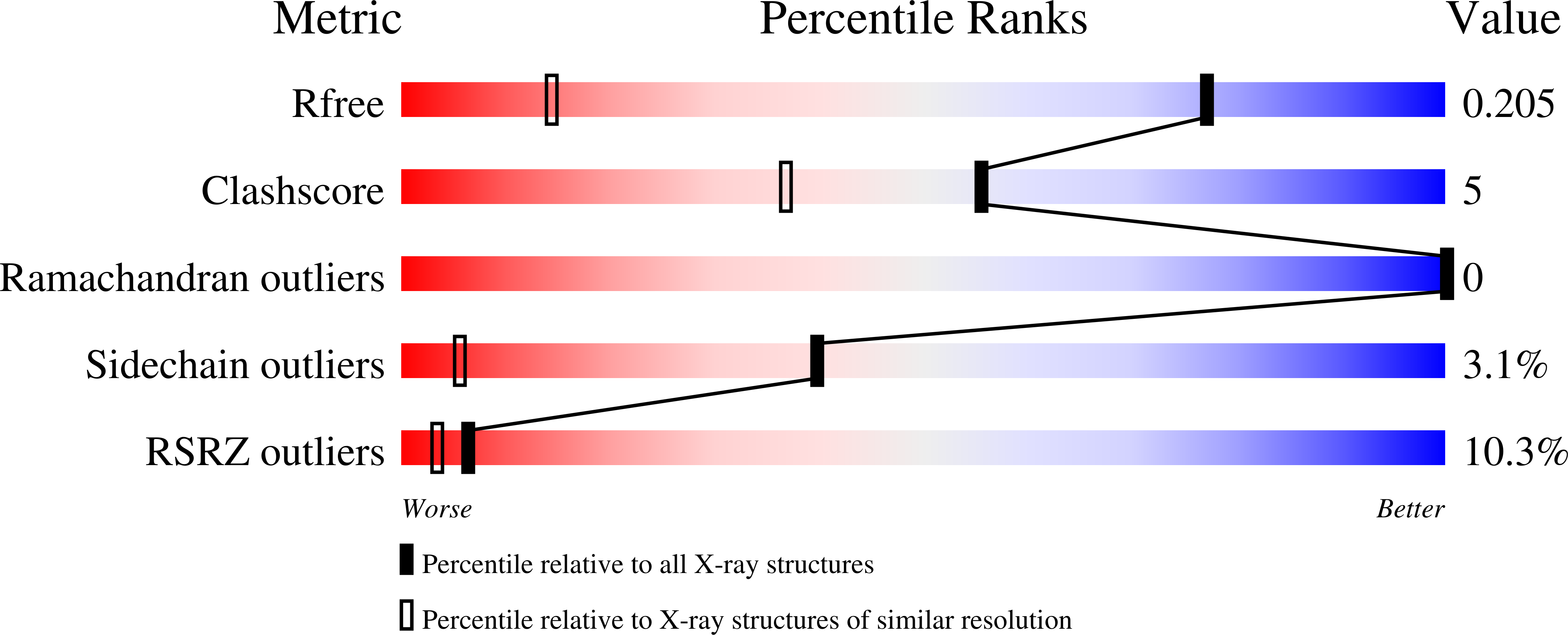
Crystallographic structure determination of protein-ligand complexes of transthyretin (TTR) has been hindered by the low affinity of many compounds that bind to the central cavity of the tetramer. Because crystallization trials are carried out at protein and ligand concentration that approach the millimolar range, low affinity is less of a problem than the poor solubility of many compounds that have been shown to inhibit amyloid fibril formation. To achieve complete occupancy in co-crystallization experiments, the minimal requirement is one ligand for each of the two sites within the TTR tetramer. Here we present a new strategy for the co-crystallization of TTR using high molecular weight polyethylene glycol instead of high ionic strength precipitants, with ligands solubilized in multicomponent mixtures of compounds. This strategy is applied to the crystallization of TTR complexes with curcumin and 16α-bromo-estradiol. Here we report the crystal structures with these compounds and with the ferulic acid that results from curcumin degradation.
Organizational Affiliation:
CEA, iBiTec-S, Service d'Ingénierie Moléculaire des Protéines, LTMB, Gif-sur-Yvette F-91191, France; Dipartimento di Farmacia, Università di Pisa, Via Bonanno 6, 56126 Pisa, Italy.