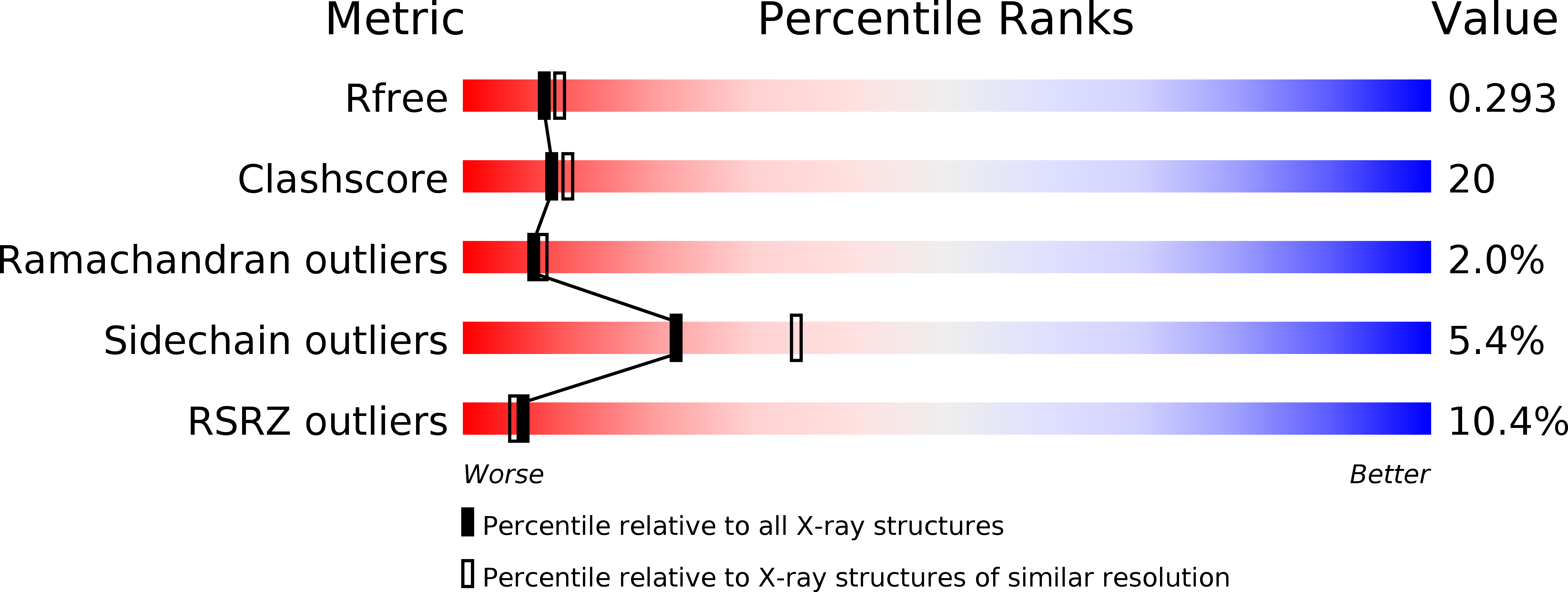
Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Singh, P.K., Sirohi, H.V., Iqbal, N., Tiwari, P., Kaur, P., Sharma, S., Singh, T.P.(2017) Biochim Biophys Acta 1865: 329-335
- PubMed: 27986533
- DOI: https://doi.org/10.1016/j.bbapap.2016.12.006
- Primary Citation of Related Structures:
2QPK, 3TGY, 4PNX, 5B72, 5GLS - PubMed Abstract:
Lactoperoxidase (LPO) is a member of mammalian heme peroxidase superfamily whose other members are myeloperoxidase (MPO), eosinophil peroxidase (EPO) and thyroid peroxidase (TPO). In these enzymes, the heme moiety is linked to protein through two or three covalent bonds. In the mature LPO, the heme moiety is linked to protein through two ester bonds with highly conserved glutamate and aspartate residues. The previously reported structures of LPO have confirmed the formation of two covalent linkages involving Glu258 and Asp108 with 1-methyl and 5-methyl groups of pyrrole rings A and C respectively. We report here a new form of structure of LPO where the covalent bond between Glu258 and 1-methyl group of pyrrole ring A is present only in a fraction of protein molecules. In this case, the side chain of Glu258 occupies two distinct positions, each of which has a 0.5 occupancy. In one position, it forms a normal ester covalent linkage while in the second position, the side chain of Glu258 is located in the middle of the substrate binding site on the distal heme side. In this position, the atom of the side chain of Glu258 forms several contacts with atoms of other residues and heme moiety. Out of the two observed positions of the side chain of Glu258, the former contributes to the stabilization of heme position and improved catalytic action of LPO while the latter is responsible for the reduced stability of the heme position as well as it blocks the substrate binding site.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.