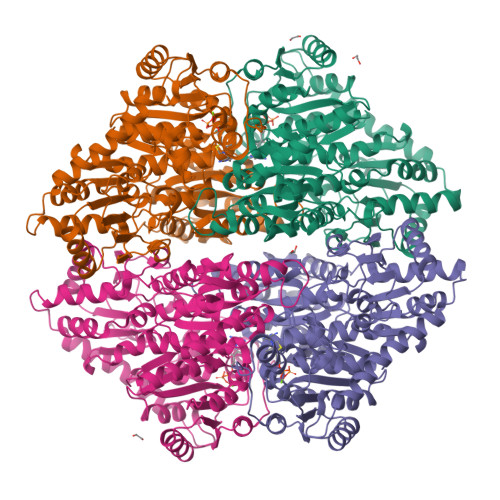
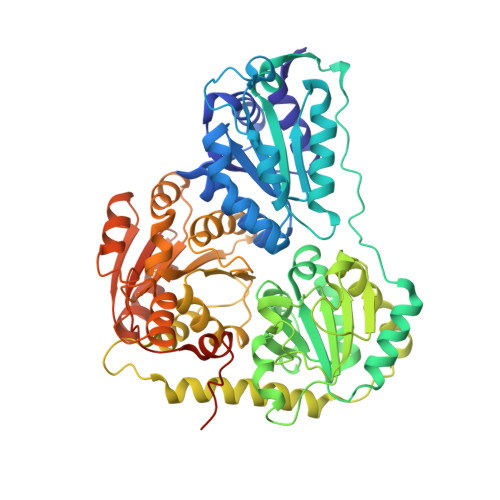
Computational protein design enables a novel one-carbon assimilation pathway.
Siegel, J.B., Smith, A.L., Poust, S., Wargacki, A.J., Bar-Even, A., Louw, C., Shen, B.W., Eiben, C.B., Tran, H.M., Noor, E., Gallaher, J.L., Bale, J., Yoshikuni, Y., Gelb, M.H., Keasling, J.D., Stoddard, B.L., Lidstrom, M.E., Baker, D.(2015) Proc Natl Acad Sci U S A 112: 3704-3709
- PubMed: 25775555
- DOI: https://doi.org/10.1073/pnas.1500545112
- Primary Citation of Related Structures:
4QPZ, 4QQ8 - PubMed Abstract:
We describe a computationally designed enzyme, formolase (FLS), which catalyzes the carboligation of three one-carbon formaldehyde molecules into one three-carbon dihydroxyacetone molecule. The existence of FLS enables the design of a new carbon fixation pathway, the formolase pathway, consisting of a small number of thermodynamically favorable chemical transformations that convert formate into a three-carbon sugar in central metabolism. The formolase pathway is predicted to use carbon more efficiently and with less backward flux than any naturally occurring one-carbon assimilation pathway. When supplemented with enzymes carrying out the other steps in the pathway, FLS converts formate into dihydroxyacetone phosphate and other central metabolites in vitro. These results demonstrate how modern protein engineering and design tools can facilitate the construction of a completely new biosynthetic pathway.
Organizational Affiliation:
Department of Chemistry, Department of Biochemistry and Molecular Medicine, and Genome Center, University of California, Davis, CA 95616; Department of Biochemistry and the Institute for Protein Design, Biomolecular Structure and Design Program.