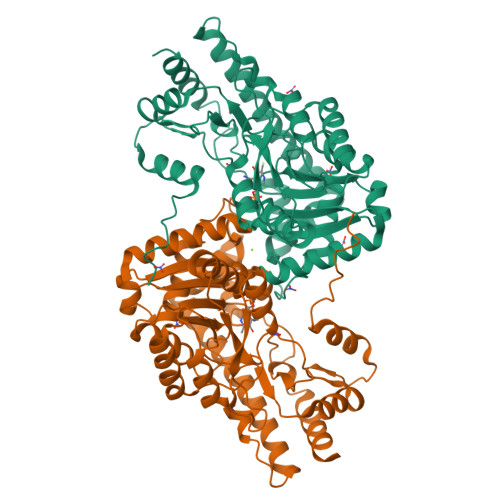
Crystal structure and enzymatic properties of a broad substrate-specificity psychrophilic aminotransferase from the Antarctic soil bacterium Psychrobacter sp. B6.
Bujacz, A., Rutkiewicz-Krotewicz, M., Nowakowska-Sapota, K., Turkiewicz, M.(2015) Acta Crystallogr D Biol Crystallogr 71: 632-645
- PubMed: 25760611
- DOI: https://doi.org/10.1107/S1399004714028016
- Primary Citation of Related Structures:
4RKC, 4RKD - PubMed Abstract:
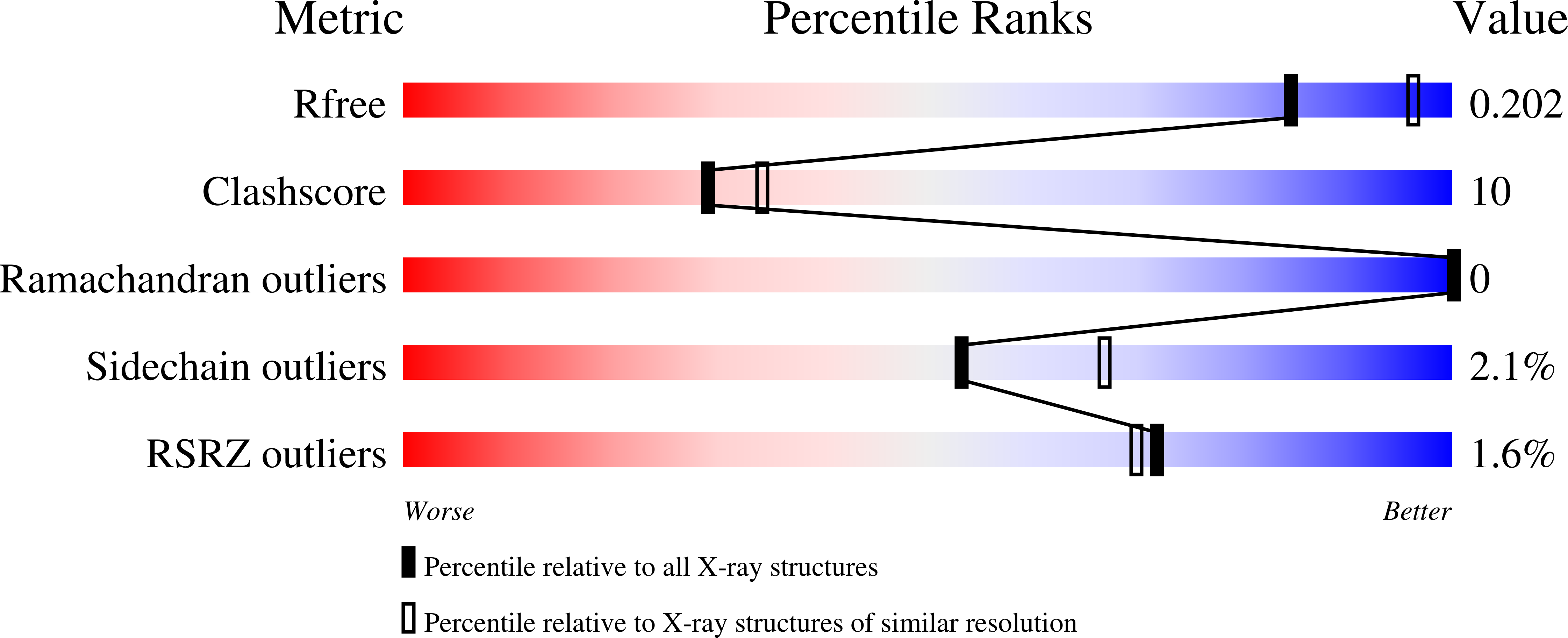
Aminotransferases (ATs) are enzymes that are commonly used in the chemical and pharmaceutical industries for the synthesis of natural and non-natural amino acids by transamination reactions. Currently, the easily accessible enzymes from mesophilic organisms are most commonly used; however, for economical and ecological reasons the utilization of aminotransferases from psychrophiles would be more advantageous, as their optimum reaction temperature is usually significantly lower than for the mesophilic ATs. Here, gene isolation, protein expression, purification, enzymatic properties and structural studies are reported for the cold-active aromatic amino-acid aminotransferase (PsyArAT) from Psychrobacter sp. B6, a psychrotrophic, Gram-negative strain from Antarctic soil. Preliminary computational analysis indicated dual functionality of the enzyme through the ability to utilize both aromatic amino acids and aspartate as substrates. This postulation was confirmed by enzymatic activity tests, which showed that it belonged to the class EC 2.6.1.57. The first crystal structures of a psychrophilic aromatic amino-acid aminotransferase have been determined at resolutions of 2.19 Å for the native enzyme (PsyArAT) and 2.76 Å for its complex with aspartic acid (PsyArAT/D). Both types of crystals grew in the monoclinic space group P21 under slightly different crystallization conditions. The PsyArAT crystals contained a dimer (90 kDa) in the asymmetric unit, which corresponds to the active form of this enzyme, whereas the crystals of the PsyArAT/D complex included four dimers showing different stages of the transamination reaction.
Organizational Affiliation:
Institute of Technical Biochemistry, Lodz University of Technology, Stefanowskiego 4/10, 90-924 Lodz, Poland.