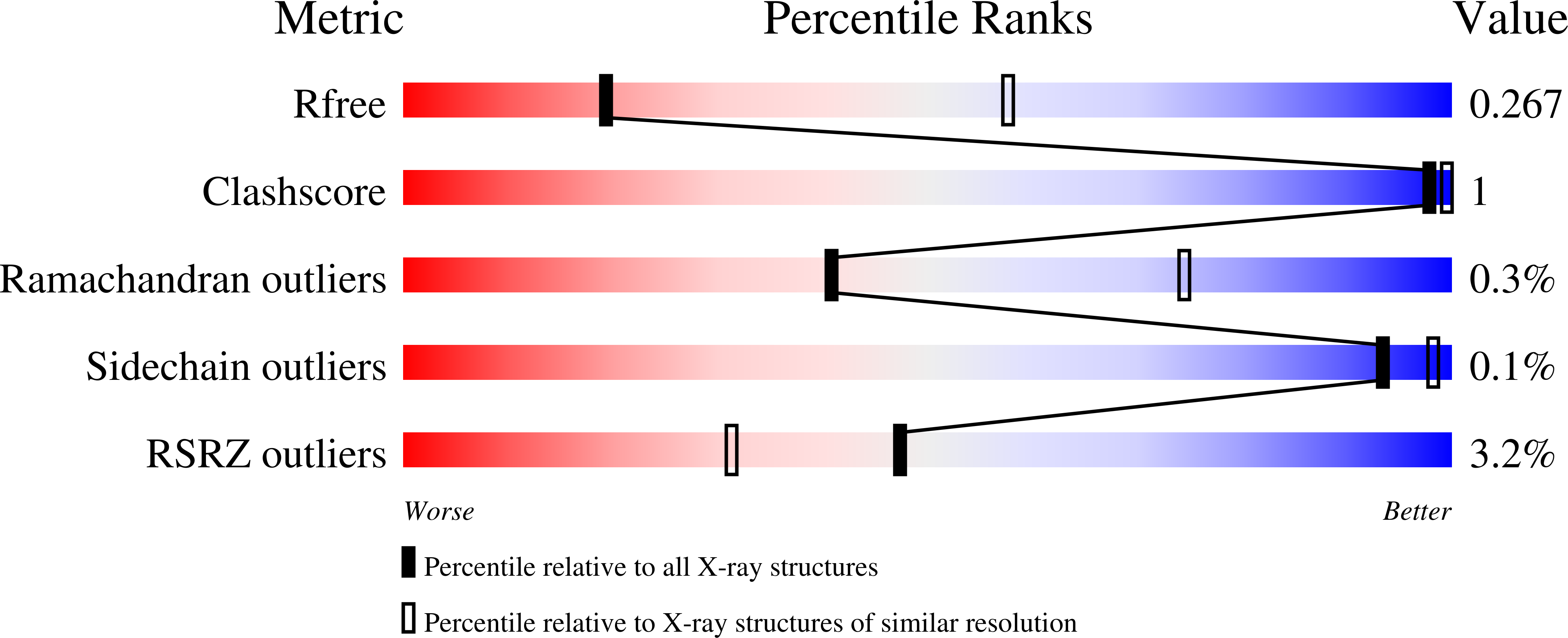
Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Bason, J.V., Montgomery, M.G., Leslie, A.G., Walker, J.E.(2014) Proc Natl Acad Sci U S A 111: 11305
- PubMed: 25049402
- DOI: https://doi.org/10.1073/pnas.1411560111
- Primary Citation of Related Structures:
4TSF, 4TT3 - PubMed Abstract:
The hydrolysis of ATP by the ATP synthase in mitochondria is inhibited by a protein called IF1. Bovine IF1 has 84 amino acids, and its N-terminal inhibitory region is intrinsically disordered. In a known structure of bovine F1-ATPase inhibited with residues 1-60 of IF1, the inhibitory region from residues 1-50 is mainly α-helical and buried deeply at the α(DP)β(DP)-catalytic interface, where it forms extensive interactions with five of the nine subunits of F1-ATPase but mainly with the β(DP)-subunit. As described here, on the basis of two structures of inhibited complexes formed in the presence of large molar excesses of residues 1-60 of IF1 and of a version of IF1 with the mutation K39A, it appears that the intrinsically disordered inhibitory region interacts first with the αEβE-catalytic interface, the most open of the three catalytic interfaces, where the available interactions with the enzyme allow it to form an α-helix from residues 31-49. Then, in response to the hydrolysis of an ATP molecule and the associated partial closure of the interface to the αTPβTP state, the extent of the folded α-helical region of IF1 increases to residues 23-50 as more interactions with the enzyme become possible. Finally, in response to the hydrolysis of a second ATP molecule and a concomitant 120° rotation of the γ-subunit, the interface closes further to the α(DP)β(DP)-state, allowing more interactions to form between the enzyme and IF1. The structure of IF1 now extends to its maximally folded state found in the previously observed inhibited complex.
Organizational Affiliation:
The Medical Research Council Mitochondrial Biology Unit, Cambridge Biomedical Campus, Cambridge CB2 0XY, United Kingdom; and.