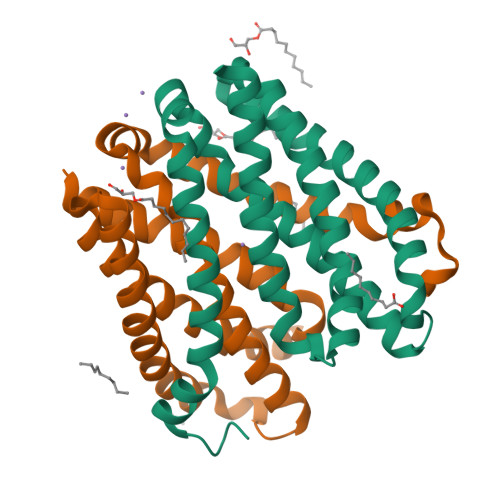
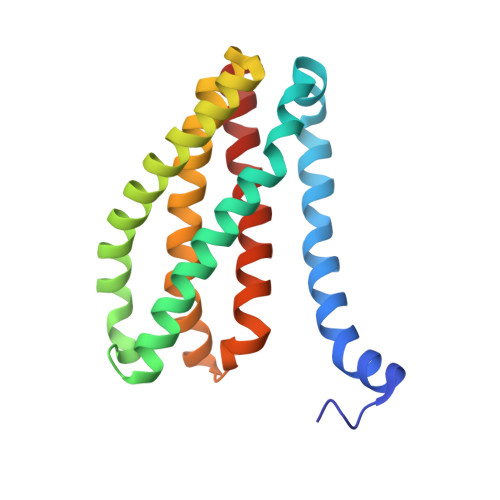
Structural basis for ion selectivity revealed by high-resolution crystal structure of Mg(2+) channel MgtE
Takeda, H., Hattori, M., Nishizawa, T., Yamashita, K., Shah, S.T., Caffrey, M., Maturana, A.D., Ishitani, R., Nureki, O.(2014) Nat Commun 5: 5374-5374
- PubMed: 25367295
- DOI: https://doi.org/10.1038/ncomms6374
- Primary Citation of Related Structures:
4U9L, 4U9N, 4WIB - PubMed Abstract:
Magnesium is the most abundant divalent cation in living cells and is crucial to several biological processes. MgtE is a Mg(2+) channel distributed in all domains of life that contributes to the maintenance of cellular Mg(2+) homeostasis. Here we report the high-resolution crystal structures of the transmembrane domain of MgtE, bound to Mg(2+), Mn(2+) and Ca(2+). The high-resolution Mg(2+)-bound crystal structure clearly visualized the hydrated Mg(2+) ion within its selectivity filter. Based on those structures and biochemical analyses, we propose a cation selectivity mechanism for MgtE in which the geometry of the hydration shell of the fully hydrated Mg(2+) ion is recognized by the side-chain carboxylate groups in the selectivity filter. This is in contrast to the K(+)-selective filter of KcsA, which recognizes a dehydrated K(+) ion. Our results further revealed a cation-binding site on the periplasmic side, which regulate channel opening and prevents conduction of near-cognate cations.
Organizational Affiliation:
1] Department of Biological Sciences, Graduate School of Science, University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan [2] Global Research Cluster, RIKEN, 2-1 Hirosawa, Wako-shi, Saitama 351-0198, Japan.