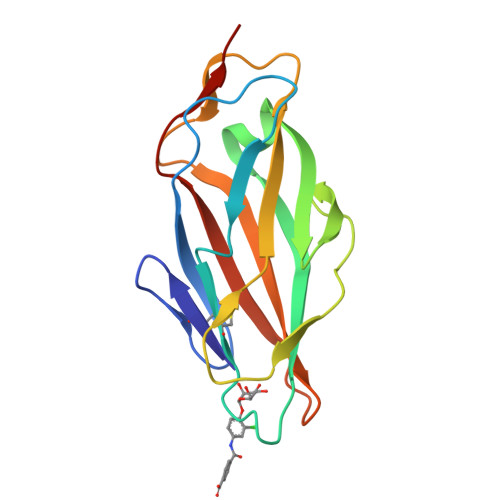
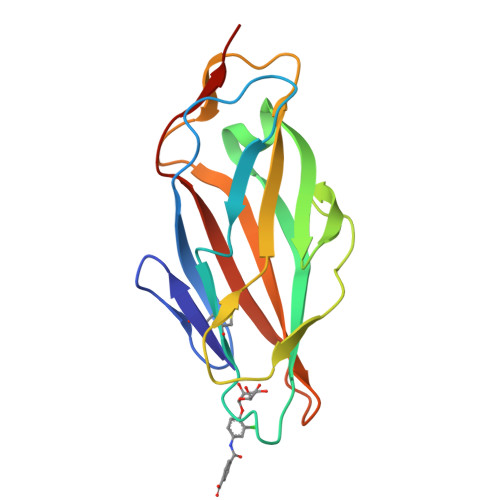
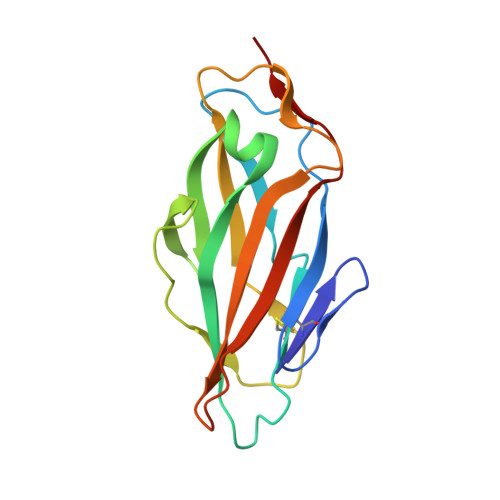
The Tyrosine Gate of the Bacterial Lectin FimH: A Conformational Analysis by NMR Spectroscopy and X-ray Crystallography.
Fiege, B., Rabbani, S., Preston, R.C., Jakob, R.P., Zihlmann, P., Schwardt, O., Jiang, X., Maier, T., Ernst, B.(2015) Chembiochem 16: 1235-1246
- PubMed: 25940742
- DOI: https://doi.org/10.1002/cbic.201402714
- Primary Citation of Related Structures:
4X50, 4X5P, 4X5Q, 4X5R - PubMed Abstract:
Urinary tract infections caused by uropathogenic E. coli are among the most prevalent infectious diseases. The mannose-specific lectin FimH mediates the adhesion of the bacteria to the urothelium, thus enabling host cell invasion and recurrent infections. An attractive alternative to antibiotic treatment is the development of FimH antagonists that mimic the physiological ligand. A large variety of candidate drugs have been developed and characterized by means of in vitro studies and animal models. Here we present the X-ray co-crystal structures of FimH with members of four antagonist classes. In three of these cases no structural data had previously been available. We used NMR spectroscopy to characterize FimH-antagonist interactions further by chemical shift perturbation. The analysis allowed a clear determination of the conformation of the tyrosine gate motif that is crucial for the interaction with aglycone moieties and was not obvious from X-ray structural data alone. Finally, ITC experiments provided insight into the thermodynamics of antagonist binding. In conjunction with the structural information from X-ray and NMR experiments the results provide a mechanism for the often-observed enthalpy-entropy compensation of FimH antagonists that plays a role in fine-tuning of the interaction.
- Institute of Molecular Pharmacy, University of Basel, Klingelbergstrasse 50, 4056 Basel (Switzerland).
Organizational Affiliation: