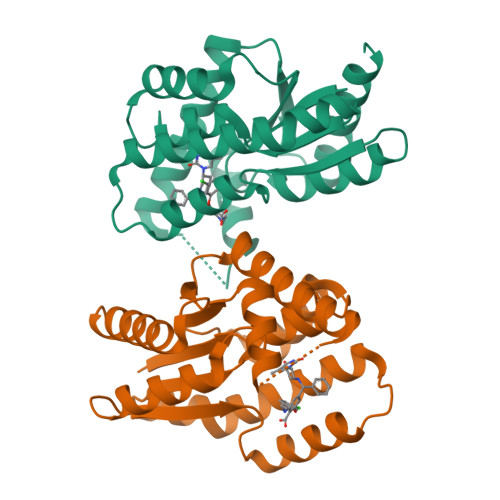
A highly potent antibacterial inhibitor of Gram-positive bacterial thymidylate kinase (TMK): SAR of piperidinyl thymines at position C5 and L1
Guler, S.Y., Martinez-Botella, G., Breen, J., Kawatkar, S., Loch, J., Olivier, N.B., Wang, H., Keating, T., Otterson, L.(2015) Bioorg Med Chem Lett