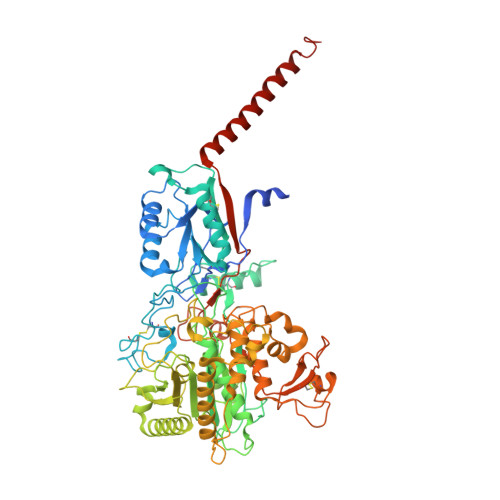
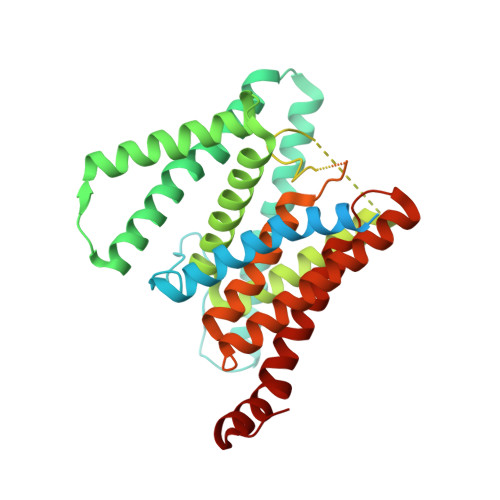
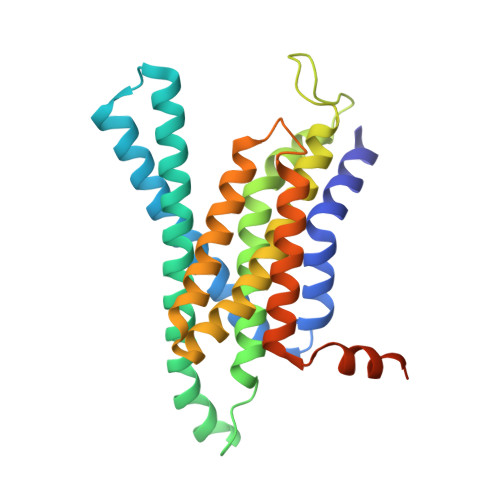
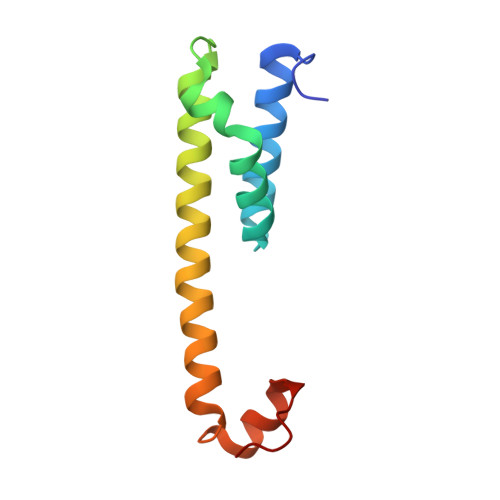
Sampling the conformational space of the catalytic subunit of human gamma-secretase.
Bai, X.C., Rajendra, E., Yang, G., Shi, Y., Scheres, S.H.(2015) Elife 4
- PubMed: 26623517
- DOI: https://doi.org/10.7554/eLife.11182
- Primary Citation of Related Structures:
5FN2, 5FN3, 5FN4, 5FN5 - PubMed Abstract:
Human γ-secretase is an intra-membrane protease that cleaves many different substrates. Aberrant cleavage of Notch is implicated in cancer, while abnormalities in cutting amyloid precursor protein lead to Alzheimer's disease. Our previous cryo-EM structure of γ-secretase revealed considerable disorder in its catalytic subunit presenilin. Here, we describe an image classification procedure that characterizes molecular plasticity at the secondary structure level, and apply this method to identify three distinct conformations in our previous sample. In one of these conformations, an additional transmembrane helix is visible that cannot be attributed to the known components of γ-secretase. In addition, we present a γ-secretase structure in complex with the dipeptidic inhibitor N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT). Our results reveal how conformational mobility in the second and sixth transmembrane helices of presenilin is greatly reduced upon binding of DAPT or the additional helix, and form the basis for a new model of how substrate enters the transmembrane domain.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge, United Kingdom.