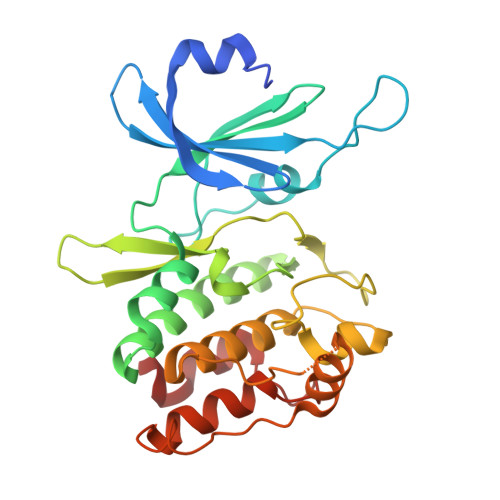
Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Patel, O., Dai, W., Mentzel, M., Griffin, M.D., Serindoux, J., Gay, Y., Fischer, S., Sterle, S., Kropp, A., Burns, C.J., Ernst, M., Buchert, M., Lucet, I.S.(2016) Structure 24: 1550-1561
- PubMed: 27545623
- DOI: https://doi.org/10.1016/j.str.2016.07.008
- Primary Citation of Related Structures:
5JZJ, 5JZN - PubMed Abstract:
Doublecortin-like kinase 1 (DCLK1) is a serine/threonine kinase that belongs to the family of microtubule-associated proteins. Originally identified for its role in neurogenesis, DCLK1 has recently been shown to regulate biological processes outside of the CNS. DCLK1 is among the 15 most common putative driver genes for gastric cancers and is highly mutated across various other human cancers. However, our present understanding of how DCLK1 dysfunction leads to tumorigenesis is limited. Here, we provide evidence that DCLK1 kinase activity negatively regulates microtubule polymerization. We present the crystal structure of the DCLK1 kinase domain at 1.7 Å resolution, providing detailed insight into the ATP-binding site that will serve as a framework for future drug design. This structure also allowed for the mapping of cancer-causing mutations within the kinase domain, suggesting that a loss of kinase function may contribute to tumorigenesis.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC 3052, Australia; Department of Medical Biology, University of Melbourne, Parkville, VIC 3052, Australia. Electronic address: patel.o@wehi.edu.au.