The Structure and Catalytic Mechanism of Sorghum bicolor Caffeoyl-CoA O-Methyltransferase.
Walker, A.M., Sattler, S.A., Regner, M., Jones, J.P., Ralph, J., Vermerris, W., Sattler, S.E., Kang, C.(2016) Plant Physiol 172: 78-92
- PubMed: 27457122
- DOI: https://doi.org/10.1104/pp.16.00845
- Primary Citation of Related Structures:
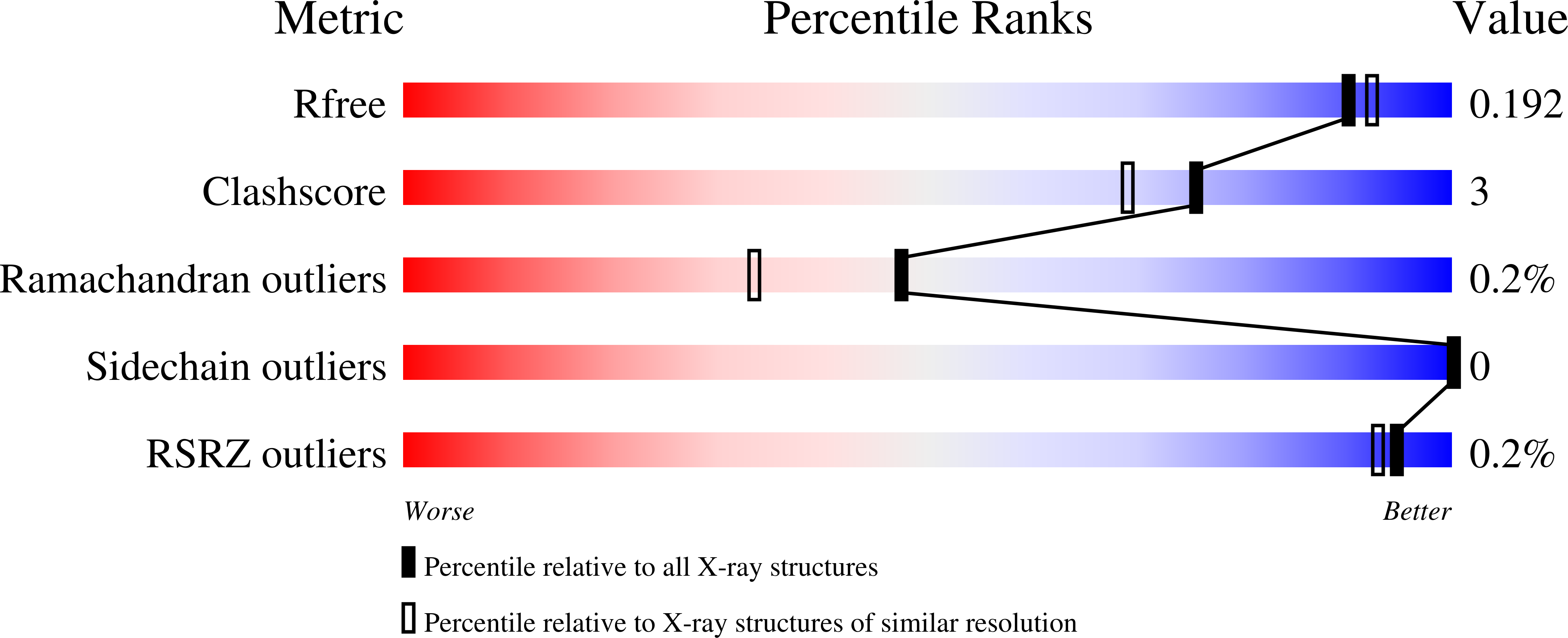
5KVA - PubMed Abstract:
Caffeoyl-coenzyme A 3-O-methyltransferase (CCoAOMT) is an S-adenosyl methionine (SAM)-dependent O-methyltransferase responsible for methylation of the meta-hydroxyl group of caffeoyl-coenzyme A (CoA) on the pathway to monolignols, with their ring methoxylation status characteristic of guaiacyl or syringyl units in lignin. In order to better understand the unique class of type 2 O-methyltransferases from monocots, we have characterized CCoAOMT from sorghum (Sorghum bicolor; SbCCoAOMT), including the SAM binary complex crystal structure and steady-state enzyme kinetics. Key amino acid residues were validated with site-directed mutagenesis. Isothermal titration calorimetry data indicated a sequential binding mechanism for SbCCoAOMT, wherein SAM binds prior to caffeoyl-CoA, and the enzyme showed allosteric behavior with respect to it. 5-Hydroxyferuloyl-CoA was not a substrate for SbCCoAOMT. We propose a catalytic mechanism in which lysine-180 acts as a catalytic base and deprotonates the reactive hydroxyl group of caffeoyl-CoA. This deprotonation is facilitated by the coordination of the reactive hydroxyl group by Ca(2+) in the active site, lowering the pKa of the 3'-OH group. Collectively, these data give a new perspective on the catalytic mechanism of CCoAOMTs and provide a basis for the functional diversity exhibited by type 2 plant OMTs that contain a unique insertion loop (residues 208-231) conferring affinity for phenylpropanoid-CoA thioesters. The structural model of SbCCoAOMT can serve as the basis for protein engineering approaches to enhance the nutritional, agronomic, and industrially relevant properties of sorghum.
Organizational Affiliation:
School of Molecular Biosciences (A.M.W., S.A.S., C.K.) and Department of Chemistry (J.P.J., C.K.), Washington State University, Pullman, Washington 99164;Department of Biochemistry and Department of Energy Great Lakes Bioenergy Research Center, University of Wisconsin, Madison, Wisconsin 53726 (M.R., J.R.);Department of Microbiology and Cell Science and Genetics Institute, University of Florida, Gainesville, Florida 32610 (W.V.); andUnited States Department of Agriculture-Agricultural Research Service, Grain Forage and Bioenergy Research Unit, Lincoln, Nebraska 68583 (S.E.S.).