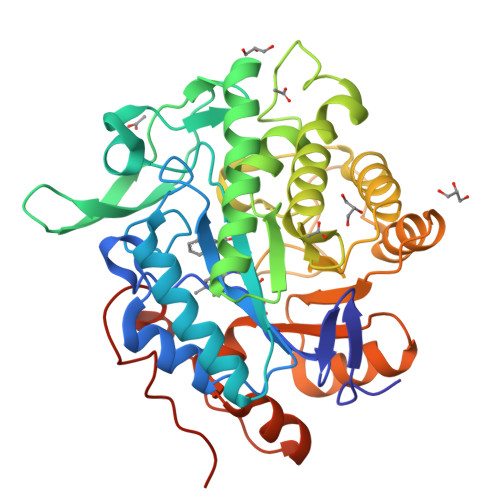
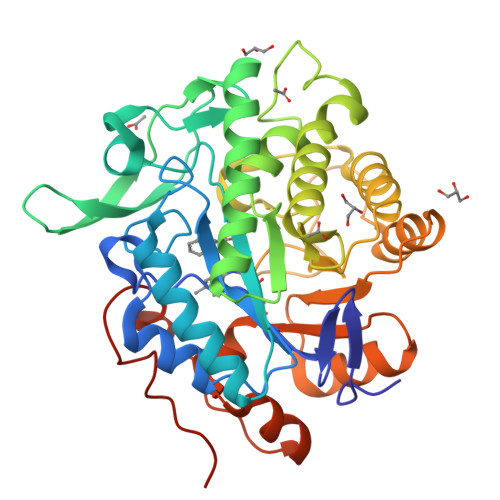
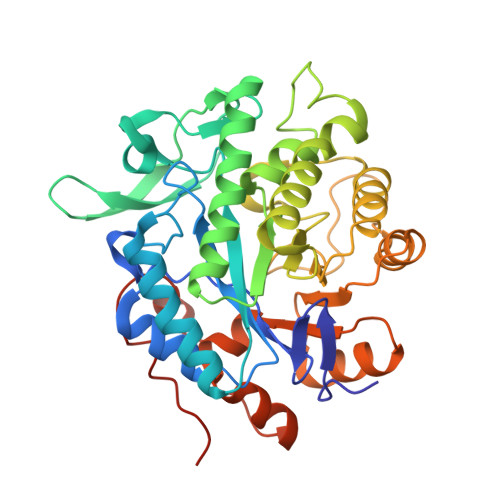
Structural insights into the ene-reductase synthesis of profens.
Waller, J., Toogood, H.S., Karuppiah, V., Rattray, N.J.W., Mansell, D.J., Leys, D., Gardiner, J.M., Fryszkowska, A., Ahmed, S.T., Bandichhor, R., Reddy, G.P., Scrutton, N.S.(2017) Org Biomol Chem 15: 4440-4448
- PubMed: 28485453
- DOI: https://doi.org/10.1039/c7ob00163k
- Primary Citation of Related Structures:
5N6G, 5N6Q - PubMed Abstract:
Reduction of double bonds of α,β-unsaturated carboxylic acids and esters by ene-reductases remains challenging and it typically requires activation by a second electron-withdrawing moiety, such as a halide or second carboxylate group. We showed that profen precursors, 2-arylpropenoic acids and their esters, were efficiently reduced by Old Yellow Enzymes (OYEs). The XenA and GYE enzymes showed activity towards acids, while a wider range of enzymes were active towards the equivalent methyl esters. Comparative co-crystal structural analysis of profen-bound OYEs highlighted key interactions important in determining substrate binding in a catalytically active conformation. The general utility of ene reductases for the synthesis of (R)-profens was established and this work will now drive future mutagenesis studies to screen for the production of pharmaceutically-active (S)-profens.
Organizational Affiliation:
Manchester Institute of Biotechnology, University of Manchester, 131 Princess Street, Manchester M1 7DN, UK. nigel.scrutton@manchester.ac.uk.