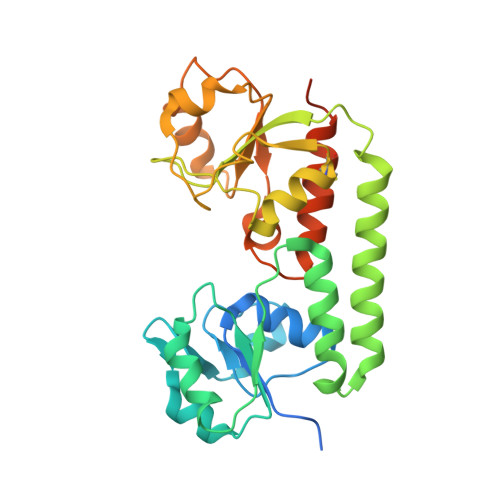
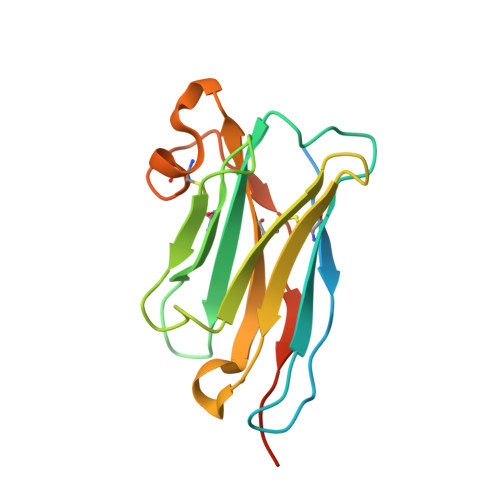
Structural basis of nanobody-mediated blocking of BtuF, the cognate substrate-binding protein of the Escherichia coli vitamin B12 transporter BtuCD.
Mireku, S.A., Sauer, M.M., Glockshuber, R., Locher, K.P.(2017) Sci Rep 7: 14296-14296
- PubMed: 29084999
- DOI: https://doi.org/10.1038/s41598-017-14512-8
- Primary Citation of Related Structures:
5OVW - PubMed Abstract:
Bacterial ABC importers catalyze the uptake of essential nutrients including transition metals and metal-containing co-factors. Recently, an IgG antibody targeting the external binding protein of the Staphylococcus aureus Mn(II) ABC importer was reported to inhibit transport activity and reduce bacterial cell growth. We here explored the possibility of using alpaca-derived nanobodies to inhibit the vitamin B12 transporter of Escherichia coli, BtuCD-F, as a model system by generating nanobodies against the periplasmic binding protein BtuF. We isolated six nanobodies that competed with B12 for binding to BtuF, with inhibition constants between 10 -6 and 10 -9 M. Kinetic characterization of the nanobody-BtuF interactions revealed dissociation half-lives between 1.6 and 6 minutes and fast association rates between 10 4 and 10 6 M -1 s -1 . For the tightest-binding nanobody, we observed a reduction of in vitro transport activity of BtuCD-F when an excess of nanobody over B12 was used. The structure of BtuF in complex with the most effective nanobody Nb9 revealed the molecular basis of its inhibitory function. The CDR3 loop of Nb9 reached into the substrate-binding pocket of BtuF, preventing both B12 binding and BtuCD-F complex formation. Our results suggest that nanobodies can mediate ABC importer inhibition, providing an opportunity for novel antibiotic strategies.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule (ETH) Zürich, CH-8093, Zürich, Switzerland.